4WFL
 
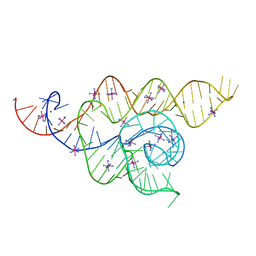 | |
4WH0
 
 | | YcaC from Pseudomonas aeruginosa with S-mercaptocysteine active site cysteine | | Descriptor: | CHLORIDE ION, Putative hydrolase | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
4WVI
 
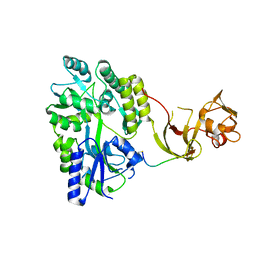 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep2). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep2) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
4WUT
 
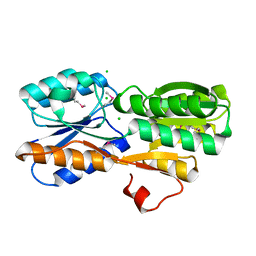 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
4X8A
 
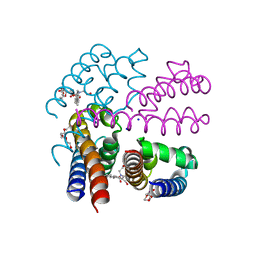 | | NavMS pore and C-terminal domain grown from protein purified in LiCl | | Descriptor: | HEGA-10, Ion transport protein, NONAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4X09
 
 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
4X91
 
 | |
4X1J
 
 | | X-ray crystal structure of the dimeric BMP antagonist NBL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Thompson, T.B, Nolan, K, Kattamuri, C. | | Deposit date: | 2014-11-24 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Neuroblastoma Suppressor of Tumorigenicity 1 (NBL1): INSIGHTS FOR THE FUNCTIONAL VARIABILITY ACROSS BONE MORPHOGENETIC PROTEIN (BMP) ANTAGONISTS.
J.Biol.Chem., 290, 2015
|
|
4WF1
 
 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4X4G
 
 | |
4WSK
 
 | | Crystal structure of a bacterial fucosidase with phenyl((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)methanone | | Descriptor: | Alpha-L-fucosidase, IMIDAZOLE, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]benzamide, ... | | Authors: | Davies, G.J. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
4WSP
 
 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-I) | | Descriptor: | CHLORIDE ION, protein DL-Rv1738 | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WSQ
 
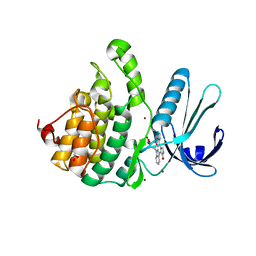 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, K-252A, ... | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Abdul, K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
4WLI
 
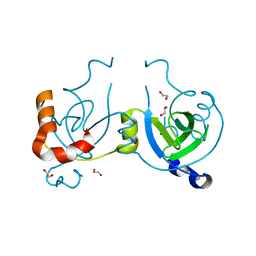 | | Stationary Phase Survival Protein YuiC from B.subtilis | | Descriptor: | 1,2-ETHANEDIOL, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLQ
 
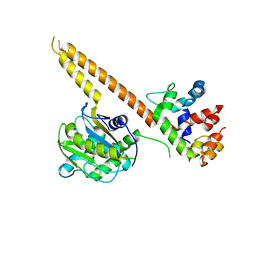 | | Crystal structure of mUCH37-hRPN13 CTD complex | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
4WT7
 
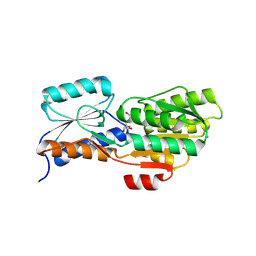 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | Descriptor: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
4WNV
 
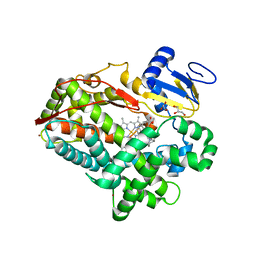 | | Human Cytochrome P450 2D6 Quinine Complex | | Descriptor: | Cytochrome P450 2D6, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
6VNZ
 
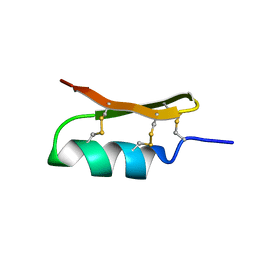 | |
4X1H
 
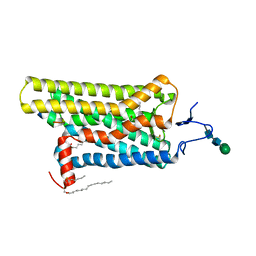 | | Opsin/G(alpha) peptide complex stabilized by nonyl-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal derived peptide of guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Blankenship, E, Lodowski, D.T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation.
Structure, 23, 2015
|
|
4X2U
 
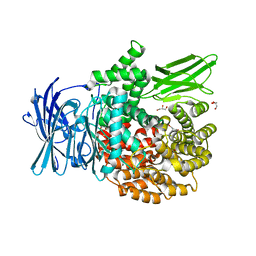 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M1 Alanyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
4X6F
 
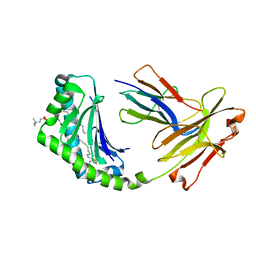 | | CD1a binary complex with sphingomyelin | | Descriptor: | (4S,7S,23Z)-4-hydroxy-7-[(1S,2Z)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-9-oxo-3,5-dioxa-8-aza-4-phosphadotriacont- 23-en-1-aminium 4-oxide, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
4X7G
 
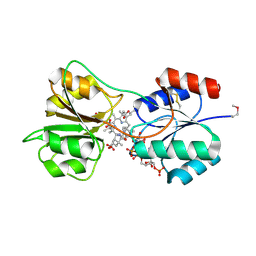 | | CobK precorrin-6A reductase | | Descriptor: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | Authors: | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
4X93
 
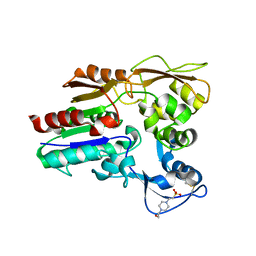 | | Crystal structure of Lysosomal Phospholipase A2 crystallized in the presence of methyl arachidonyl fluorophosphonate (tetragonal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase.
Nat Commun, 6, 2015
|
|
4WMZ
 
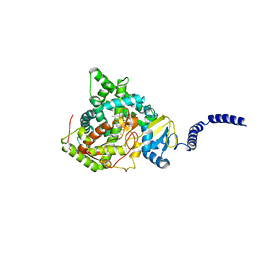 | | S. cerevisiae CYP51 complexed with fluconazole in the active site | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Wilbanks, S.M, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 59, 2015
|
|
4PI3
 
 | |
