6U23
 
 | | EM structure of MPEG-1(w.t.) soluble pre-pore | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6U2L
 
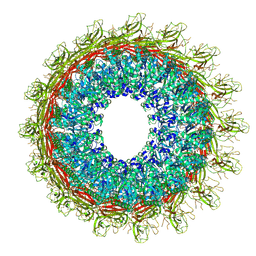 | | EM structure of MPEG-1 (L425K, beta conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6UIM
 
 | |
6U2K
 
 | |
6UIJ
 
 | |
6U7X
 
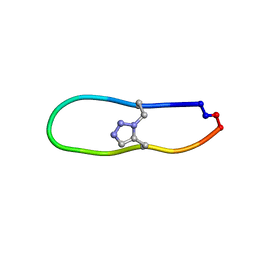 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UHU
 
 | |
1PJU
 
 | | Unbound form of Tomato Inhibitor-II | | Descriptor: | SULFATE ION, Wound-induced proteinase inhibitor II | | Authors: | Barrette-Ng, I.H, Ng, K.K.-S, Cherney, M.M, Pearce, G, Ghani, U, Ryan, C.A, James, M.N.G. | | Deposit date: | 2003-06-03 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unbound form of tomato inhibitor-II reveals interdomain flexibility and conformational variability in the reactive site loops
J.Biol.Chem., 278, 2003
|
|
3SC2
 
 | | REFINED ATOMIC MODEL OF WHEAT SERINE CARBOXYPEPTIDASE II AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE II (CPDW-II), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liao, D.-I, Remington, S.J. | | Deposit date: | 1992-07-01 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined atomic model of wheat serine carboxypeptidase II at 2.2-A resolution.
Biochemistry, 31, 1992
|
|
1H6K
 
 | | nuclear Cap Binding Complex | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, CBP80 | | Authors: | Mazza, C, Ohno, M, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Nuclear CAP Binding Complex
Mol.Cell, 8, 2001
|
|
3TC2
 
 | |
1K09
 
 | | Solution structure of BetaCore, A Designed Water Soluble Four-Stranded Antiparallel b-sheet Protein | | Descriptor: | (7E)-4,9-dioxo-6-oxa-3,7,10-triazadodec-7-ene-1,12-dioic acid, Core Module I, Core Module II | | Authors: | Carulla, N, Woodward, C, Barany, G. | | Deposit date: | 2001-09-18 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-21 | | Method: | SOLUTION NMR | | Cite: | BetaCore, a designed water soluble four-stranded antiparallel beta-sheet protein.
Protein Sci., 11, 2002
|
|
3U0S
 
 | | Crystal Structure of an Enzyme Redesigned Through Multiplayer Online Gaming: CE6 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diisopropyl-fluorophosphatase, GLYCEROL, ... | | Authors: | Bale, J.B, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.
Nat.Biotechnol., 30, 2012
|
|
3W1O
 
 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
2GGV
 
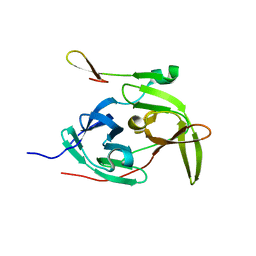 | | Crystal structure of the West Nile virus NS2B-NS3 protease, His51Ala mutant | | Descriptor: | non-structural protein 2B, non-structural protein 3 | | Authors: | Aleshin, A.E, Shiryaev, S.A, Strongin, A.Y, Liddington, R.C. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold.
Protein Sci., 16, 2007
|
|
2GJ0
 
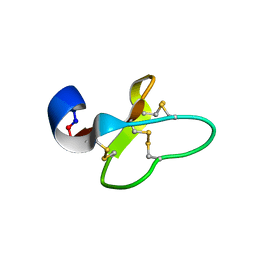 | | Cycloviolacin O14 | | Descriptor: | Cycloviolacin O14 | | Authors: | Ireland, D.C, Colgrave, M.L, Craik, D.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-04-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A novel suite of cyclotides from Viola odorata: sequence variation and the implications for structure, function and stability
Biochem.J., 400, 2006
|
|
2RNG
 
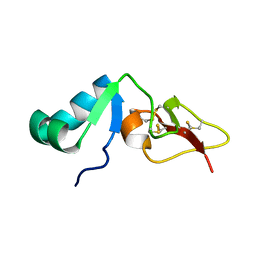 | | Solution structure of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Fujitani, N, Osaki, T, Kawabata, S, Nishimura, S, Mizuguchi, M, Aizawa, T, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Biochemistry, 47, 2008
|
|
2RQY
 
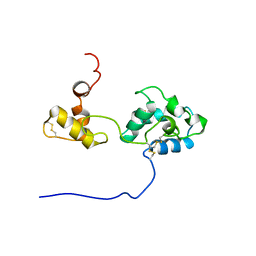 | | Solution structure and dynamics of mouse ARMET | | Descriptor: | Putative uncharacterized protein | | Authors: | Hoseki, J, Sasakawa, H, Yamaguchi, Y, Maeda, M, Kubota, H, Kato, K, Nagata, K. | | Deposit date: | 2010-01-26 | | Release date: | 2010-04-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of mouse ARMET.
Febs Lett., 584, 2010
|
|
2UUX
 
 | | Structure of the tryptase inhibitor TdPI from a tick | | Descriptor: | SULFATE ION, TRYPTASE INHIBITOR | | Authors: | Siebold, C, Paesen, G.C, Harlos, K, Peacey, M.F, Nuttall, P.A, Stuart, D.I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
J.Mol.Biol., 368, 2007
|
|
2V3T
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form | | Descriptor: | CALCIUM ION, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT SYNONYM GLURDELTA2, GLUR DELTA-2 | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2QN4
 
 | | Structure and function study of rice bifunctional alpha-amylase/subtilisin inhibitor from Oryza sativa | | Descriptor: | Alpha-amylase/subtilisin inhibitor | | Authors: | Peng, W.Y, Lin, Y.H, Huang, Y.C, Guan, H.H, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function Study of Rice Bifunctional Alpha-Amylase/Subtilisin Inhibitor from Oryza Sativa
To be Published
|
|
2V3U
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | Descriptor: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | Authors: | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-06-22 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2FBW
 
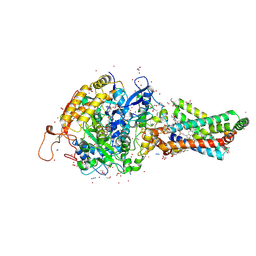 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
2I83
 
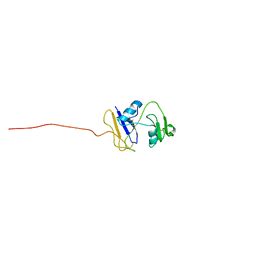 | | hyaluronan-binding domain of CD44 in its ligand-bound form | | Descriptor: | CD44 antigen | | Authors: | Takeda, M, Ogino, S, Umemoto, R, Sakakura, M, Kajiwara, M, Sugahara, K.N, Hayasaka, H, Miyasaka, M, Terasawa, H, Shimada, I. | | Deposit date: | 2006-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Ligand-induced Structural Changes of the CD44 Hyaluronan-binding Domain Revealed by NMR
J.Biol.Chem., 281, 2006
|
|
2REL
 
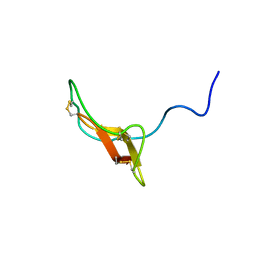 | | SOLUTION STRUCTURE OF R-ELAFIN, A SPECIFIC INHIBITOR OF ELASTASE, NMR, 11 STRUCTURES | | Descriptor: | R-ELAFIN | | Authors: | Francart, C, Dauchez, M, Alix, A.J.P, Lippens, G. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of R-elafin, a specific inhibitor of elastase.
J.Mol.Biol., 268, 1997
|
|
