9BKM
 
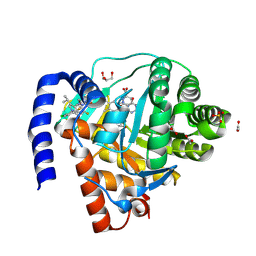 | | DHODH in complex with Ligand 10 | | Descriptor: | (2M,6P)-2-(2-chloro-6-fluorophenyl)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-4-(propan-2-yl)isoquinolin-1(2H)-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 67, 2024
|
|
9BKD
 
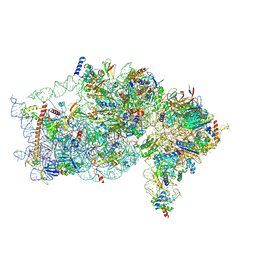 | | The structure of human Pdcd4 bound to the 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Albacete-Albacete, L, Ramakrishnan, V, S.Fraser, C. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
9BK3
 
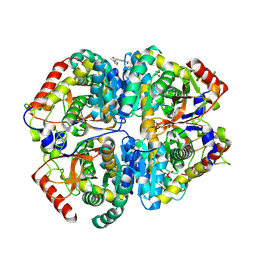 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BK2
 
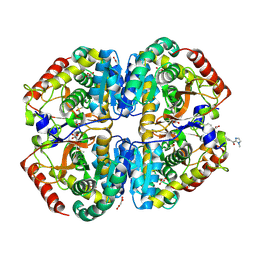 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BJ1
 
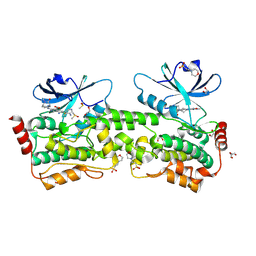 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
9BIK
 
 | | Crystal structure of inhibitor 1 bound to HPK1 | | Descriptor: | (1S,2S)-N-[(6P)-8-amino-6-(4-methylpyridin-3-yl)isoquinolin-3-yl]-2-cyanocyclopropane-1-carboxamide, 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Kiefer, J.T, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Siu, M, Heffron, T.P, Choo, E.F. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
9BIG
 
 | | Stat6 bound to degrader AK-1690 | | Descriptor: | Signal transducer and activator of transcription 6, [(2-{[(2S)-1-{(2S,4S)-4-[(7-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}hept-6-yn-1-yl)oxy]-2-[(2R)-2-phenylmorpholine-4-carbonyl]pyrrolidin-1-yl}-3,3-dimethyl-1-oxobutan-2-yl]carbamoyl}-1-benzothiophen-5-yl)di(fluoro)methyl]phosphonic acid | | Authors: | Mallik, L, Stuckey, J.A. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Discovery of AK-1690: A Potent and Highly Selective STAT6 PROTAC Degrader.
J.Med.Chem., 2024
|
|
9BI8
 
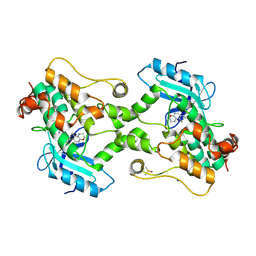 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (3R,4S)-4-methyloxolan-3-yl [(6P)-8-amino-7-fluoro-6-(8-methyl-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)isoquinolin-3-yl]carbamate, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase kinase 1, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-22 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
9BHK
 
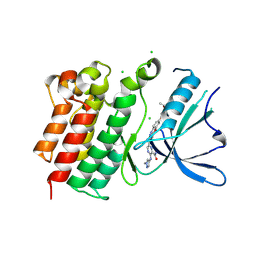 | | MerTK in complex with small molecule inhibitor 6-{1-[6-(3-hydroxy-3-methylbutoxy)-1,3-benzoxazol-2-yl]azetidin-3-yl}-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide | | Descriptor: | 6-{1-[6-(3-hydroxy-3-methylbutoxy)-1,3-benzoxazol-2-yl]azetidin-3-yl}-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | Jakob, C.G, Gurbani, D, Qiu, W. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Discovery of Potent Azetidine-Benzoxazole MerTK Inhibitors with In Vivo Target Engagement.
J.Med.Chem., 2024
|
|
9BHJ
 
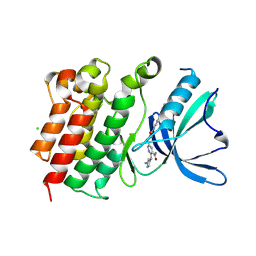 | |
9BHI
 
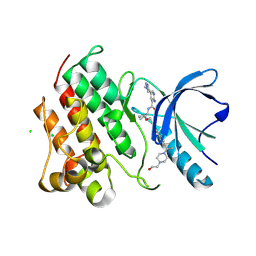 | | Crystal structure of the MerTK kinase domain with SA4488 | | Descriptor: | (5P)-2-amino-5-(1-methyl-1H-pyrazol-4-yl)-N-{(1R,2S)-2-[(4'-{2-[4-(2-oxoethyl)piperazin-1-yl]propan-2-yl}[1,1'-biphenyl]-4-yl)methoxy]cyclopentyl}pyridine-3-carboxamide, CHLORIDE ION, Mer tyrosine kinase domain | | Authors: | Jakob, C.G, Qui, W, Jain, R. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of A-910, a Highly Potent and Orally Bioavailable Dual MerTK/Axl-Selective Tyrosine Kinase Inhibitor.
J.Med.Chem., 2024
|
|
9BH5
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | High-Resolution Reconstruction of a C. elegans Ribosome Sheds Light on Evolutionary Dynamics and Tissue Specificity.
Rna, 2024
|
|
9BGE
 
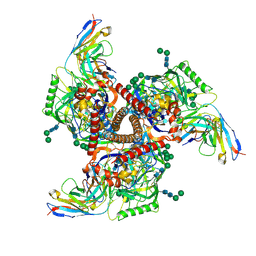 | | Cryo-EM structure of mAb8-24 bound to 426c.WITO.TM.SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c.WITO.TM.SOSIP, ... | | Authors: | Hurlburt, N.K, Pancera, M. | | Deposit date: | 2024-04-18 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Short CDRL1 in intermediate VRC01-like mAbs is not sufficient to overcome key glycan barriers on HIV-1 Env.
J.Virol., 2024
|
|
9BFY
 
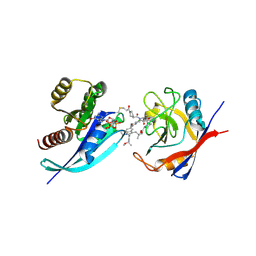 | | Tri-complex of Compound-12, KRAS G12C, and CypA | | Descriptor: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Tri-complex of Compound-12, KRAS G12C, and CypA
To be published
|
|
9BFT
 
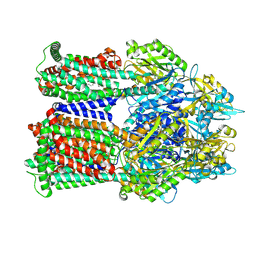 | | Cryo-EM co-structure of AcrB with CU244 | | Descriptor: | (2S)-1-{[(1R,5R)-3-azabicyclo[3.1.0]hexan-6-yl]amino}-3-(3,5-dichlorophenoxy)propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFN
 
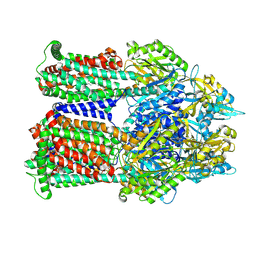 | | Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor | | Descriptor: | (2R)-1-(4-aminopiperidin-1-yl)-3-[3-(trifluoromethyl)phenoxy]propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFM
 
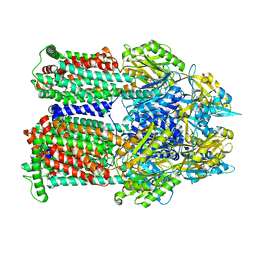 | | Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor | | Descriptor: | (2S)-1-(3,4-dichlorophenoxy)-3-(4-{[4-(trifluoromethyl)pyrimidin-2-yl]amino}piperidin-1-yl)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFH
 
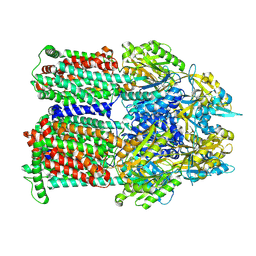 | | Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor | | Descriptor: | (2S)-1-[(3R)-3-aminopyrrolidin-1-yl]-3-(3,4-dichlorophenoxy)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFG
 
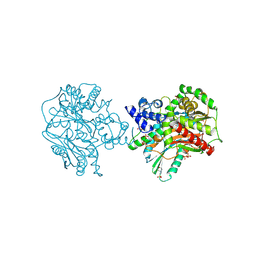 | | Structure of the crosslinked PCP-E didomain of tyrocidine synthetase A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-pentyl-beta-alaninamide, ... | | Authors: | Heberlig, G.W, Burkart, M.D. | | Deposit date: | 2024-04-17 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crosslinking Reveals the Molecular Details of Intermodular Condensation and Epimerization in Nonribosomal Peptide Biosynthesis
To Be Published
|
|
9BFB
 
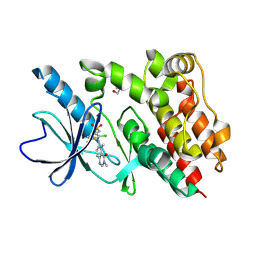 | | Crystal structure of BRAF kinase domain with PF-07284890 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{2-chloro-3-[(3,5-dimethyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-fluorophenyl}-3-fluoropropane-1-sulfonamide, ... | | Authors: | Mou, T.-C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of the Clinical Candidate PF-07284890 ( ARRY-461 ), a Highly Potent and Brain Penetrant BRAF Inhibitor for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
9BF6
 
 | |
9BEW
 
 | |
9BER
 
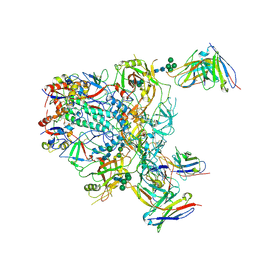 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
9BEQ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with AP1867 | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-04-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PROTAC-mediated activation, rather than degradation, of a nuclear receptor reveals complex ligand-receptor interaction network
Structure, 2024
|
|
9BEI
 
 | |
