3DHC
 
 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
7LEV
 
 | |
5Z9K
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-[(2R)-2-(hydroxymethyl)pyrrolidin-1-yl]sulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5Z2C
 
 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
2BHP
 
 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-01-16 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
7LNH
 
 | | S-adenosylmethionine synthetase co-crystallized with UppNHp | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-$l^{3}-sulfanyl]butanoic acid, 1,2-ETHANEDIOL, S-adenosylmethionine synthase isoform type-2 | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
3LC7
 
 | | Crystal Structure of apo Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from methicllin resistant Staphylococcus aureus (MRSA252) | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1 | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3ES4
 
 | |
4CLR
 
 | |
4CMC
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4-cyclohexyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CMJ
 
 | |
4CME
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4,N4-dimethyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CLH
 
 | |
4CJX
 
 | | The crystal structure of Trypanosoma brucei N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, ... | | Authors: | Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-12-23 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of 2,4-Diamino-6-Oxo-1,6-Dihydropyrimidin-5-Yl Ureido Based Inhibitors of Trypanosoma Brucei Fold and Testing for Antiparasitic Activity.
J.Med.Chem., 58, 2015
|
|
4GCM
 
 | |
4CMB
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | ACETATE ION, N4-cyclohexyl-5-(4-fluorophenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
6UDJ
 
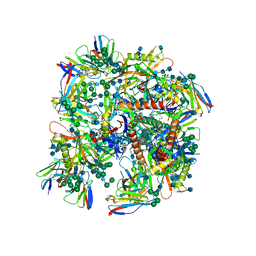 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UDK
 
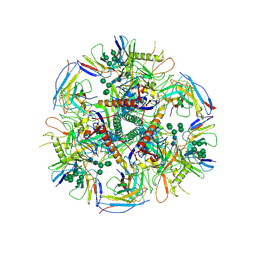 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
3NO5
 
 | |
3O76
 
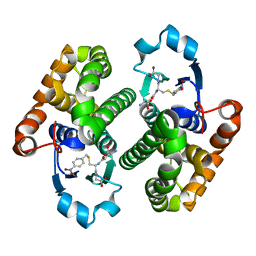 | |
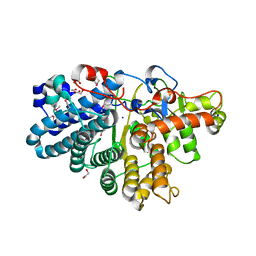
3IHV
 
 | |
6H5W
 
 | |
3ANN
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with quinolin-2-ylmethylphosphonic acid | | Descriptor: | (quinolin-2-ylmethyl)phosphonic acid, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Endo, K, Kato, M, Deng, L, Song, Y, Yajima, S. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes
ACS Med Chem Lett, 2, 2011
|
|
1VOK
 
 | | ARABIDOPSIS THALIANA TBP (DIMER) | | Descriptor: | SULFATE ION, TATA-BOX-BINDING PROTEIN | | Authors: | Nikolov, D.B, Burley, S.K. | | Deposit date: | 1996-04-29 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 A resolution refined structure of a TATA box-binding protein (TBP).
Nat.Struct.Biol., 1, 1994
|
|
3ANR
 
 | | human DYRK1A/harmine complex | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
