7U23
 
 | |
1I3C
 
 | | RESPONSE REGULATOR FOR CYANOBACTERIAL PHYTOCHROME, RCP1 | | Descriptor: | RESPONSE REGULATOR RCP1, SULFATE ION | | Authors: | Im, Y.J, Rho, S.-H, Park, C.-M, Yang, S.-S, Kang, J.-G, Lee, J.Y, Song, P.-S, Eom, S.H. | | Deposit date: | 2001-02-14 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a cyanobacterial phytochrome response regulator.
Protein Sci., 11, 2002
|
|
4IXE
 
 | | pcDHFR-NADPH-270 | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(3,4,5-trifluorophenyl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Design, Synthesis, and Molecular Modeling of Novel Pyrido[2,3-d]pyrimidine Analogues As Antifolates; Application of Buchwald-Hartwig Aminations of Heterocycles.
J.Med.Chem., 56, 2013
|
|
8ZXQ
 
 | | Crystal structure of Ssr1698 in complex with heme b | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Ssr1698 protein | | Authors: | Wang, X, Liu, L. | | Deposit date: | 2024-06-14 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis for Monomer-Dimer Transition of Dri1 Upon Heme Binding.
Proteins, 93, 2025
|
|
4IRM
 
 | |
4IXG
 
 | | pcDHFR-268-K37S-N69F variant | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-phenylpyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Molecular Modeling of Novel Pyrido[2,3-d]pyrimidine Analogues As Antifolates; Application of Buchwald-Hartwig Aminations of Heterocycles.
J.Med.Chem., 56, 2013
|
|
4IXF
 
 | | pcDHFR-269 F69N variant | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-[4-(propan-2-yl)phenyl]pyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Molecular Modeling of Novel Pyrido[2,3-d]pyrimidine Analogues As Antifolates; Application of Buchwald-Hartwig Aminations of Heterocycles.
J.Med.Chem., 56, 2013
|
|
2A2F
 
 | | Crystal Structure of Sec15 C-terminal domain | | Descriptor: | Exocyst complex component Sec15 | | Authors: | Wu, S, Mehta, S.Q, Pichaud, F, Bellen, H.J, Quiocho, F.A. | | Deposit date: | 2005-06-22 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2ZVT
 
 | | Cys285Ser mutant PPARgamma ligand-binding domain complexed with 15-deoxy-delta12,14-prostaglandin J2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Oyama, T, Shiraki, T, Morikawa, K. | | Deposit date: | 2008-11-19 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure of mutant PPARgamma LBD complexed with 15d-PGJ2: novel modulation mechanism of PPARgamma/RXRalpha function by covalently bound ligands
Febs Lett., 583, 2009
|
|
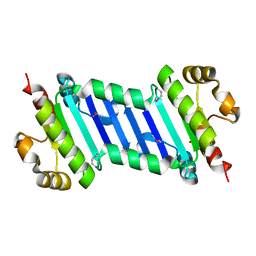
6KI2
 
 | |
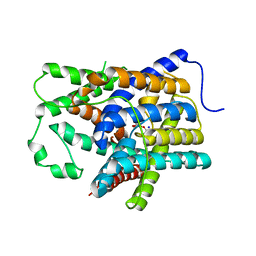
6KI1
 
 | |
6L8D
 
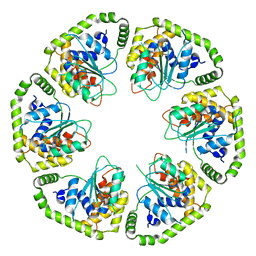 | |
5U0B
 
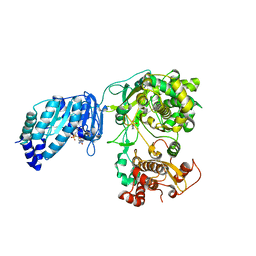 | | Structure of full-length Zika virus NS5 | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhao, B, Du, F. | | Deposit date: | 2016-11-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the Zika virus full-length NS5 protein.
Nat Commun, 8, 2017
|
|
3Q17
 
 | | Structure of a slow CLC Cl-/H+ antiporter from a cyanobacterium in Bromide | | Descriptor: | BROMIDE ION, Sll0855 protein | | Authors: | Jayaram, H, Robertson, J.L, Fang, W, Williams, C, Miller, C. | | Deposit date: | 2010-12-16 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of a Slow CLC Cl(-)/H(+) Antiporter from a Cyanobacterium.
Biochemistry, 50, 2011
|
|
6HAG
 
 | |
8AY2
 
 | |
4YNL
 
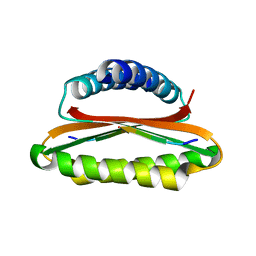 | | Crystal structure of the hood domain of Anabaena HetR in complex with the hexapeptide ERGSGR derived from PatS | | Descriptor: | Heterocyst differentiation control protein, Heterocyst inhibition-signaling peptide | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Zhang, C.C, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-03-10 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into HetR-PatS interaction involved in cyanobacterial pattern formation
Sci Rep, 5, 2015
|
|
3TE3
 
 | |
3E5E
 
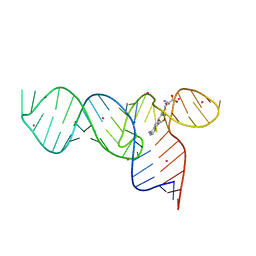 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2PWY
 
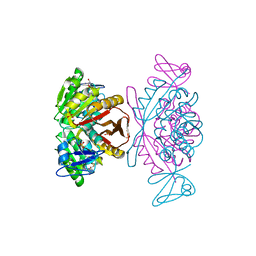 | | Crystal Structure of a m1A58 tRNA methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (adenine-N(1)-)-methyltransferase | | Authors: | Barraud, P, Golinelli-Pimpaneau, B, Tisne, C. | | Deposit date: | 2007-05-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermus thermophilus tRNA m(1)A(58) Methyltransferase and Biophysical Characterization of Its Interaction with tRNA.
J.Mol.Biol., 377, 2008
|
|
2YGI
 
 | | Methanobactin HM1 | | Descriptor: | COPPER (II) ION, METHANOBACTIN HM1 | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YGJ
 
 | | Methanobactin MB4 | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB4, SODIUM ION | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3DN9
 
 | |
3OSJ
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 254-400) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | NITRATE ION, Phycobilisome LCM core-membrane linker polypeptide, SODIUM ION | | Authors: | Kuzin, A, Su, M, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-09 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209C
To be published
|
|
3OHW
 
 | | X-Ray Structure of Phycobilisome LCM core-membrane linker polypeptide (fragment 721-860) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR209E | | Descriptor: | Phycobilisome LCM core-membrane linker polypeptide | | Authors: | Kuzin, A, Su, M, Lew, S, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR209E
To be Published
|
|
