5NSL
 
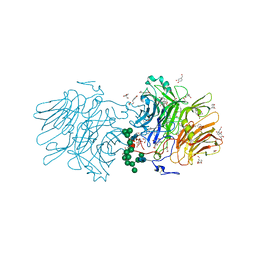 | |
4TLV
 
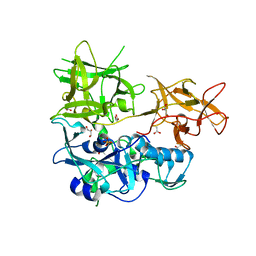 | | CARDS TOXIN, NICKED | | Descriptor: | ACETATE ION, ADP-ribosylating toxin CARDS, GLYCEROL, ... | | Authors: | Taylor, A.B, Pakhomova, O.N, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5LZX
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a UGA stop codon. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
4TNK
 
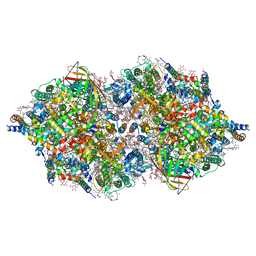 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4ZP3
 
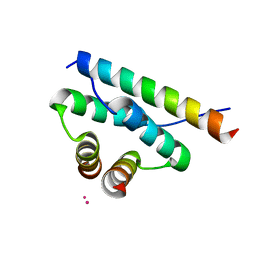 | | AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA | | Descriptor: | A-kinase anchor protein 7 isoforms alpha and beta, CADMIUM ION, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Goetz, F, Roske, Y, Faelber, K, Zuehlke, K, Autenrieth, K, Kreuchwig, A, Krause, G, Herberg, F.W, Daumke, O, Heinemann, U, Klussmann, E. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | AKAP18:PKA-RII alpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.
Biochem.J., 473, 2016
|
|
5FYJ
 
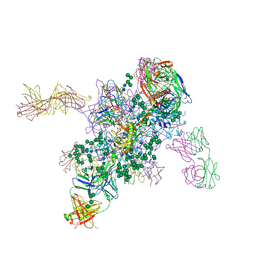 | | Crystal Structure at 3.4 A Resolution of Fully Glycosylated HIV-1 Clade G X1193.c1 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
4TOE
 
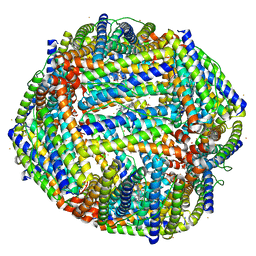 | | 2.20A resolution structure of Iron Bound BfrB (D34F) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4ZKH
 
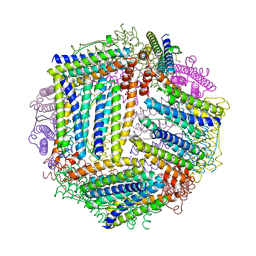 | |
5LTV
 
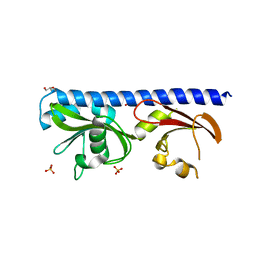 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5E4J
 
 | | Acetylcholinesterase Methylene Blue no PEG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, DECAMETHONIUM ION, ... | | Authors: | Dym, O. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The impact of crystallization conditions on structure-based drug design: A case study on the methylene blue/acetylcholinesterase complex.
Protein Sci., 25, 2016
|
|
6CK4
 
 | | G96A mutant of the PRPP riboswitch from T. mathranii bound to ppGpp | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Reiss, C.W, Knappenberger, A.J, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
6GSK
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and near-cognate tRNAThr in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria.
Nucleic Acids Res., 46, 2018
|
|
5E23
 
 | | Human transthyretin (TTR) complexed with (2,7-Dibromo-fluoren-9-ylideneaminooxy)-acetic acid | | Descriptor: | DIMETHYL SULFOXIDE, Transthyretin, {[(2,7-dibromo-9H-fluoren-9-ylidene)amino]oxy}acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR).
J Enzyme Inhib Med Chem, 31, 2016
|
|
5E2B
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
4TSY
 
 | | Crystal structure of FraC with lipids | | Descriptor: | 2-[[(E,2S,3R)-2-(hexanoylamino)-3-oxidanyl-dec-4-enoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, Fragaceatoxin C, HEPTANE-1,2,3-TRIOL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
6GYB
 
 | | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri | | Descriptor: | VirB10 protein, VirB7, VirB9 protein | | Authors: | Sgro, G.G, Costa, T.R.D, Farah, C.S, Waksman, G. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri.
Nat Microbiol, 3, 2018
|
|
6SP8
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 prepared by the 'soak-and-freeze' method under 150 bar of krypton pressure | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
6H01
 
 | |
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
6CNL
 
 | | Crystal Structure of H105A PGAM5 Dodecamer | | Descriptor: | MAGNESIUM ION, PGAM5 Multimerization Motif Peptide, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
6H23
 
 | | Crystal structure of the hClpP Y118A mutant with an activating small molecule | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Kick, L.M, Sieber, S.A, Schneider, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Selective Activation of Human Caseinolytic Protease P (ClpP).
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5LZS
 
 | | Structure of the mammalian ribosomal elongation complex with aminoacyl-tRNA, eEF1A, and didemnin B | | Descriptor: | (2~{S})-~{N}-[(2~{R})-1-[[(3~{S},6~{S},8~{S},12~{S},13~{R},16~{S},17~{R},20~{S},23~{S})-13-[(2~{S})-butan-2-yl]-20-[(4-methoxyphenyl)methyl]-6,17,21-trimethyl-3-(2-methylpropyl)-12-oxidanyl-2,5,7,10,15,19,22-heptakis(oxidanylidene)-8-propan-2-yl-9,18-dioxa-1,4,14,21-tetrazabicyclo[21.3.0]hexacosan-16-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-~{N}-methyl-1-[(2~{S})-2-oxidanylpropanoyl]pyrrolidine-2-carboxamide, 18S ribosomal RNA, 28S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZB
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5M0I
 
 | | Crystal structure of the nuclear complex with She2p and the ASH1 mRNA E3-localization element | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASH1-E3 element, ... | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4RSZ
 
 | |
