2DIR
 
 | | Solution structure of the THUMP domain of THUMP domain-containing protein 1 | | Descriptor: | THUMP domain-containing protein 1 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the THUMP domain of THUMP domain-containing protein 1
To be Published
|
|
2Q15
 
 | | Structure of BACE complexed to compound 3a | | Descriptor: | (4S)-4-(2-AMINO-6-PHENOXYQUINAZOLIN-3(4H)-YL)-N,4-DICYCLOHEXYL-N-METHYLBUTANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
4B4L
 
 | | CRYSTAL STRUCTURE OF AN ARD DAP-KINASE 1 MUTANT | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, PENTAETHYLENE GLYCOL, SULFATE ION | | Authors: | Temmerman, K, Pogenberg, V, Jonko, W, Wilmanns, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Pef/Y Substrate Recognition and Signature Motif Plays a Critical Role in Dapk-Related Kinase Activity.
Chem.Biol., 21, 2014
|
|
3VUK
 
 | | Crystal structure of a cysteine-deficient mutant M5 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-07-02 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
2AAY
 
 | | EPSP synthase liganded with shikimate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, ... | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A. | | Deposit date: | 2005-07-14 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
4FLS
 
 | | Crystal structure of Amylosucrase inactive double mutant F290K-E328Q from Neisseria polysaccharea in complex with sucrose. | | Descriptor: | Amylosucrase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
3VUL
 
 | | Crystal structure of a cysteine-deficient mutant M1 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1 | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-07-02 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
3R20
 
 | |
2Q5D
 
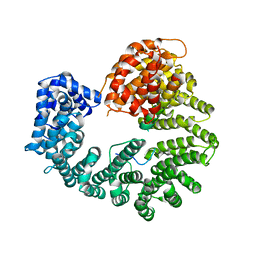 | |
4OPR
 
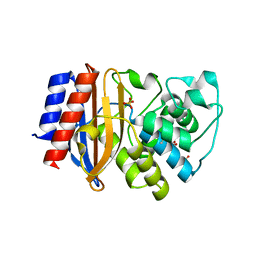 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-06 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
2W1U
 
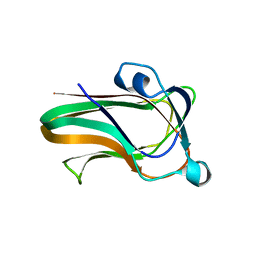 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ACETATE ION, ... | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2PU1
 
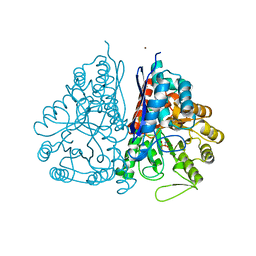 | | Crystal Structure of the T. brucei enolase complexed with Fluoro-phosphonoacetohydroxamate (FPAH) | | Descriptor: | 1,2-ETHANEDIOL, Enolase, ZINC ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
3EWH
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridyl-pyrimidine benzimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-6-(trifluoromethyl)-1H-benzimidazol-2-amine, vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3MB8
 
 | | Crystal structure of purine nucleoside phosphorylase from toxoplasma gondii in complex with immucillin-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ho, M, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition and Structure of Toxoplasma gondii Purine Nucleoside Phosphorylase.
Eukaryot Cell, 13, 2014
|
|
2DE3
 
 | | Crystal structure of DSZB C27S mutant in complex with 2'-hydroxybiphenyl-2-sulfinic acid | | Descriptor: | 2'-HYDROXY-1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, ... | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
5QH8
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with PCM-0102558 | | Descriptor: | 1-[(2S)-2-(4-methoxyphenyl)-2,3,6,7-tetrahydro-1H-azepin-1-yl]ethan-1-one, ACETATE ION, Peroxisomal coenzyme A diphosphatase NUDT7 | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
6QP7
 
 | | Drosophila Semaphorin 2a | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
1IN2
 
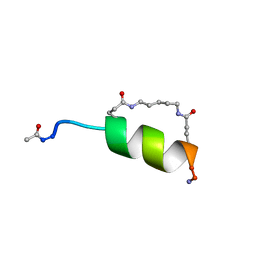 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
5QJ9
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z768399682 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4GY9
 
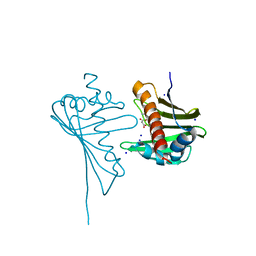 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5QK7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1918536193 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-4-[(1-methyl-1H-pyrazol-4-yl)oxy]pyrimidine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJM
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z328695024 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3DPS
 
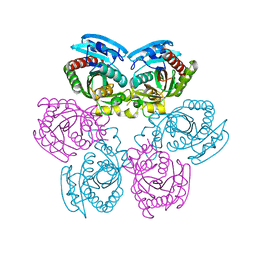 | |
2PX2
 
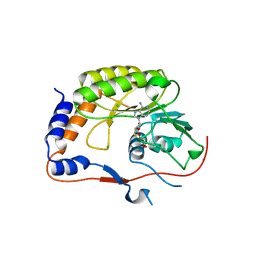 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Monoclinic form 1) | | Descriptor: | CHLORIDE ION, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
3PX3
 
 | | Structure of TylM1 from Streptomyces fradiae H123A mutant in complex with SAH and dTDP-Quip3N | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Holden, H.M, Carney, A.E. | | Deposit date: | 2010-12-09 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Architecture of TylM1 from Streptomyces fradiae: An N,N-Dimethyltransferase Involved in the Production of dTDP-d-mycaminose .
Biochemistry, 50, 2011
|
|
