6T58
 
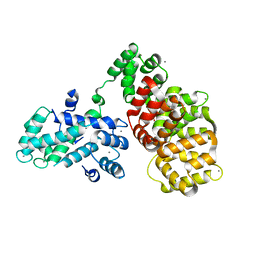 | | Structure determination of the transactivation domain of p53 in complex with S100A4 using annexin A2 as a crystallization chaperone | | Descriptor: | CALCIUM ION, Cellular tumor antigen p53,Protein S100-A4,Protein S100-A4,Annexin A2, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Nyitray, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure Determination of the Transactivation Domain of p53 in Complex with S100A4 Using Annexin A2 as a Crystallization Chaperone.
Structure, 28, 2020
|
|
8RJC
 
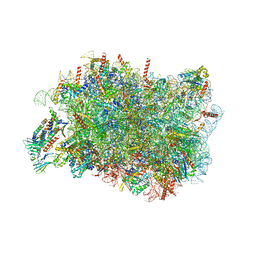 | |
8RJD
 
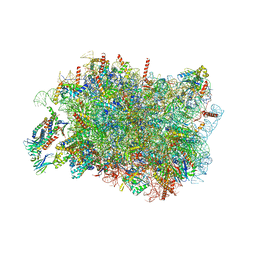 | |
8G8E
 
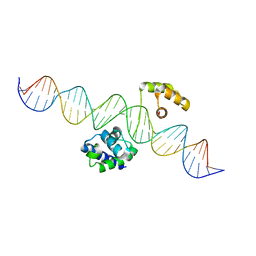 | | Human Oct4 bound to nucleosome with human LIN28B sequence | | Descriptor: | LIN28B DNA (32-MER), POU domain, class 5, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8YD7
 
 | |
8YD8
 
 | |
8YBX
 
 | |
6YBD
 
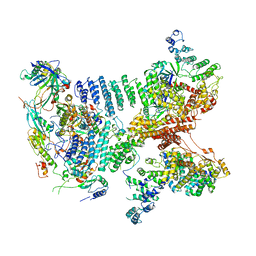 | | Structure of a human 48S translational initiation complex - eIF3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14, 40S ribosomal protein S17, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
8G87
 
 | | Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of Oct4 bound region) | | Descriptor: | POU domain, class 5, transcription factor 1, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
7WU0
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B3 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTX
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTZ
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B2 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
1YFD
 
 | | Crystal structure of the Y122H mutant of ribonucleotide reductase R2 protein from E. coli | | Descriptor: | MERCURY (II) ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Kolberg, M, Logan, D.T, Bleifuss, G, Poetsch, S, Sjoeberg, B.M, Graeslund, A, Lubitz, W, Lassmann, G, Lendzian, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies
J.Biol.Chem., 280, 2005
|
|
6C23
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C24
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
4LYN
 
 | | Crystal structure of cyclin-dependent kinase 2 (cdk2-wt) complex with (2s)-n-(5-(((5-tert-butyl-1,3-oxazol-2-yl)methyl)sulfanyl)-1,3-thiazol-2-yl)-2-phenylpropanamide | | Descriptor: | (2S)-N-(5-{[(5-tert-butyl-1,3-oxazol-2-yl)methyl]sulfanyl}-1,3-thiazol-2-yl)-2-phenylpropanamide, Cyclin-dependent kinase 2 | | Authors: | Sack, J.S. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminothiazole Inhibitors of Cyclin-Dependent Kinase 2: Synthesis, X-Ray Crystallographic Analysis, and Biological Activities
J.Med.Chem., 45, 2002
|
|
5IFE
 
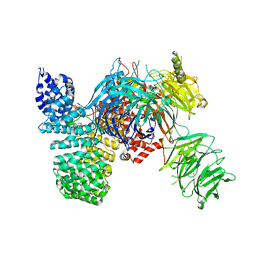 | | Crystal structure of the human SF3b core complex | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Cretu, C, Dybkov, O, De Laurentiis, E, Will, C.L, Luhrmann, R, Pena, V. | | Deposit date: | 2016-02-25 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations.
Mol.Cell, 64, 2016
|
|
7YI1
 
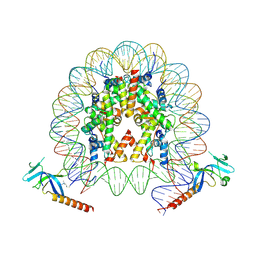 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
6PY8
 
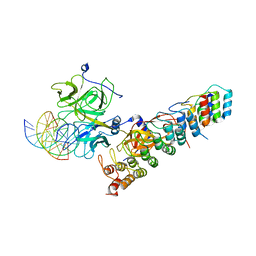 | | Crystal structure of the RBPJ-NOTCH1-NRARP ternary complex bound to DNA | | Descriptor: | DNA, Neurogenic locus notch homolog protein 1, Notch-regulated ankyrin repeat-containing protein, ... | | Authors: | Jarrett, S.M, Seegar, T.C.M, Blacklow, S.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Sci.Signal., 12, 2019
|
|
7YI5
 
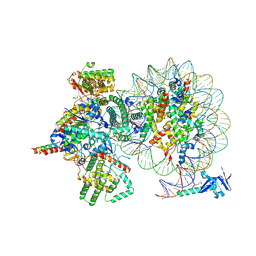 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI4
 
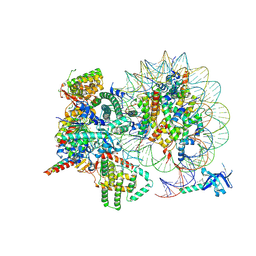 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7PMK
 
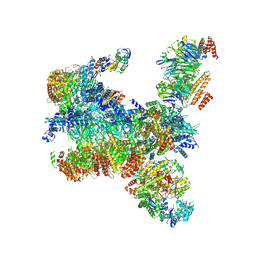 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation I) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA helicase, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
8T12
 
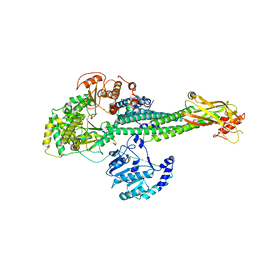 | |
8T13
 
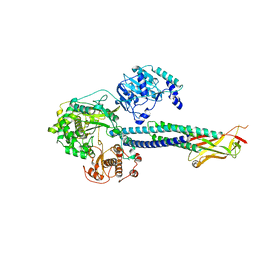 | |
