3SYT
 
 | | Crystal structure of glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi, NAD+, and glutamate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Chuenchor, W, Gerratana, B. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6511 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
1V8S
 
 | | Crystal structure analusis of the ADP-ribose pyrophosphatase complexed with AMP and Mg | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
3O7L
 
 | |
3TY9
 
 | | Crystal Structure of C. Thermocellum PNKP Ligase Domain AMP-Adenylate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Smith, P, Wang, L, Shuman, S. | | Deposit date: | 2011-09-24 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2BZK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PIM1 IN COMPLEX WITH AMPPNP AND PIMTIDE | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PIMTIDE, PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1 | | Authors: | Debreczeni, J.E, Bullock, A, Knapp, S, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-08-18 | | Release date: | 2005-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human Pim1 in Complex with Amppnp and Pimtide
To be Published
|
|
6BKG
 
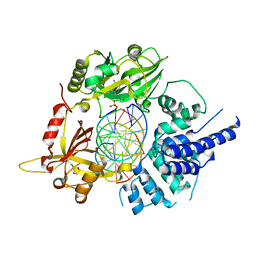 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
1J1Z
 
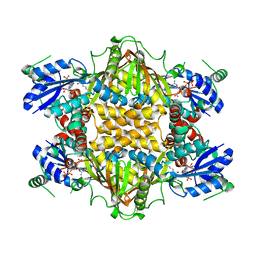 | |
5WS9
 
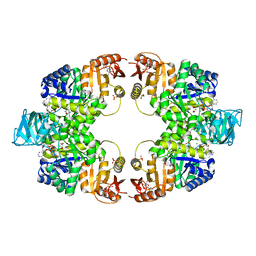 | | Pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, ATP and allosteric activator AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
3A7A
 
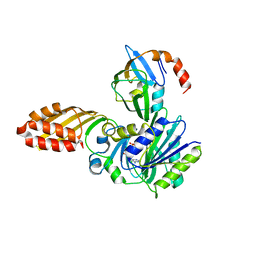 | | Crystal structure of E. coli lipoate-protein ligase A in complex with octyl-amp and apoH-protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine cleavage system H protein, Lipoate-protein ligase A, ... | | Authors: | Fujiwara, K, Hosaka, H, Nakagawa, A. | | Deposit date: | 2009-09-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
2GMK
 
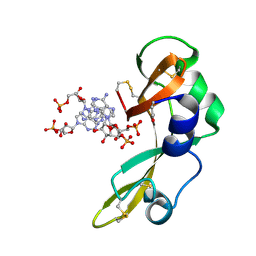 | | Crystal structure of onconase double mutant with spontaneously-assembled (AMP) 4 stack | | Descriptor: | ADENOSINE MONOPHOSPHATE, P-30 protein | | Authors: | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
5GZW
 
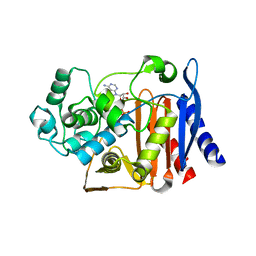 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
3A7R
 
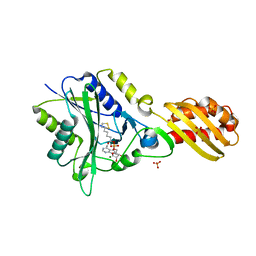 | |
2QVU
 
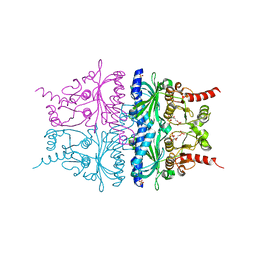 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Mg2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
2QVV
 
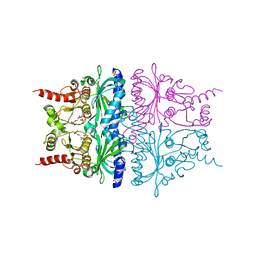 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Zn2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, PHOSPHATE ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
6AIB
 
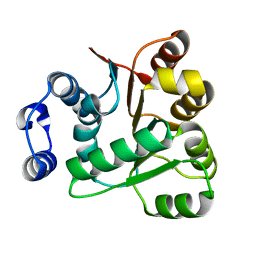 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4A73
 
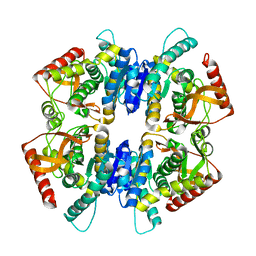 | |
3ZZN
 
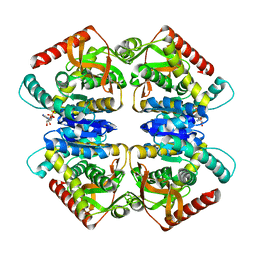 | | 5-Mutant (R79W, R151A, E279A, E299A,E313A) Lactate-Dehydrogenase from Thermus thermophillus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, LACTATE DEHYDROGENASE | | Authors: | Colletier, J.P, Mraihi, S, Madern, D. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sampling the conformational energy landscape of a hyperthermophilic protein by engineering key substitutions.
Mol. Biol. Evol., 29, 2012
|
|
2QVR
 
 | | E. coli Fructose-1,6-bisphosphatase: Citrate, Fru-2,6-P2, and Mg2+ bound | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CITRIC ACID, Fructose-1,6-bisphosphatase, ... | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of Mammalian and Bacterial Fructose-1,6-bisphosphatase Reveal the Basis for Synergism in AMP/Fructose 2,6-Bisphosphate Inhibition.
J.Biol.Chem., 282, 2007
|
|
6A06
 
 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
2RMK
 
 | | Rac1/PRK1 Complex | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Modha, R, Campbell, L.J, Nietlispach, D, Buhecha, H.R, Owen, D, Mott, H.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Rac1 Polybasic Region Is Required for Interaction with Its Effector PRK1
J.Biol.Chem., 283, 2008
|
|
2DSD
 
 | |
2ECK
 
 | | STRUCTURE OF PHOSPHOTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE | | Authors: | Berry, M.B, Bilderback, T, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP/AMP complex of Escherichia coli adenylate kinase.
Proteins, 62, 2006
|
|
6FH2
 
 | | Protein arginine kinase McsB in the AMP-PN-bound state | | Descriptor: | 1,2-ETHANEDIOL, AMP PHOSPHORAMIDATE, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6GB0
 
 | |
6GBC
 
 | |
