5L2O
 
 | | Crystal Structure of ALDH1A1 in complex with BUC22 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Buchman, C.D, Hurley, T.D. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives.
J. Med. Chem., 60, 2017
|
|
3FPY
 
 | | Azurin C112D/M121L | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Lancaster, K.M, Gray, H.B. | | Deposit date: | 2009-01-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Zero Copper Proteins.
Nat Chem, 1, 2009
|
|
9QM5
 
 | |
7KQ9
 
 | |
3D9F
 
 | | Nitroalkane oxidase: active site mutant S276A crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3DDD
 
 | |
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
8TJC
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 8a | | Descriptor: | (6M)-1-[(2R)-3,3-dimethylbutan-2-yl]-6-[(5S)-5-methyl-4-oxo-5-phenyl-4,5-dihydro-1H-imidazol-2-yl]-1,3-dihydro-2H-benzimidazol-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-20 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
3D9E
 
 | | Nitroalkane oxidase: active site mutant D402N crystallized with 1-nitrooctane | | Descriptor: | 1-nitrooctane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3D9D
 
 | | Nitroalkane oxidase: mutant D402N crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
5IZC
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor F032 | | Descriptor: | ACETATE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pozzi, C, Landi, G, Di Pisa, F, Mangani, S. | | Deposit date: | 2016-03-25 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
7A9H
 
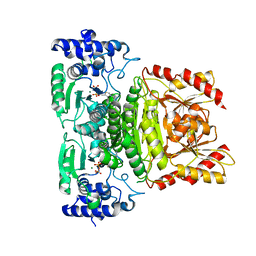 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
9FYZ
 
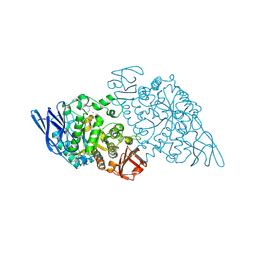 | | Crystal structure of SusA amylase from Bacteroides thetaiotaomicron covalently bound to alpha-1,6 branched pseudo-trisaccharide activity-based probe | | Descriptor: | (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Pickles, I.B, Moroz, O, Davies, G. | | Deposit date: | 2024-07-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Precision Activity-Based alpha-Amylase Probes for Dissection and Annotation of Linear and Branched-Chain Starch-Degrading Enzymes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FZ2
 
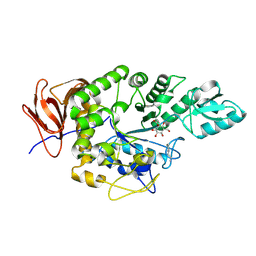 | | Crystal structure of Amylase 5 (Amy5) from Ruminococcus bromii covalently bound to alpha-1,6 branched pseudo-trisaccharide activity-based probe | | Descriptor: | (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 8-azanyloctan-1-ol, Alpha-amylase, ... | | Authors: | Pickles, I.B, Moroz, O, Davies, G. | | Deposit date: | 2024-07-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Precision Activity-Based alpha-Amylase Probes for Dissection and Annotation of Linear and Branched-Chain Starch-Degrading Enzymes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
3EIF
 
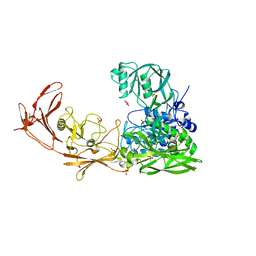 | | 1.9 angstrom crystal structure of the active form of the C5a peptidase from Streptococcus pyogenes (ScpA) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Cooney, J.C, Kagawa, T.F, O'Connell, M.R, Paoli, M, Mouat, P, O'Toole, P.W. | | Deposit date: | 2008-09-15 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 A Crystal Structure of the Active Form of ScpA
J.Mol.Biol., 386, 2009
|
|
6H5I
 
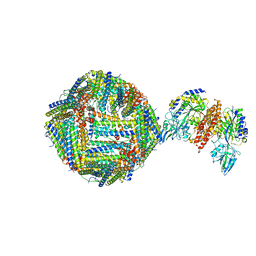 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P, Savino, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
6H9B
 
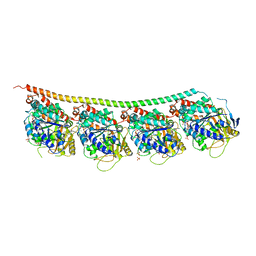 | | 1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation | | Descriptor: | 9-methyl-3-[1-(2-methylquinolin-4-yl)ethenyl]carbazole, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2018-08-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin-Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation.
J. Med. Chem., 62, 2019
|
|
6NN4
 
 | | The structure of human liver pyruvate kinase, hLPYK-D499N, in complex with Fru-1,6-BP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, PHOSPHOENOLPYRUVATE, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7MX6
 
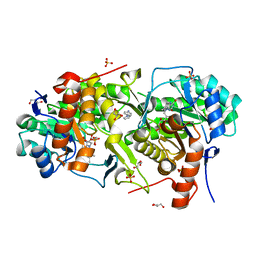 | | Leishmania major dihydroorotate dehydrogenase in complex with [4-(1H-pyrrol-1-yl)phenyl]methanol | | Descriptor: | 4-(1H-pyrrol-1-yl)aniline, Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Hunter, W.N, Cardoso, I.A, Nonato, M.C. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Leishmania major dihydroorotate dehydrogenase in complex with [4-(1H-pyrrol-1-yl)phenyl]methanol
To be Published
|
|
7MYD
 
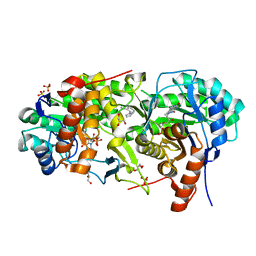 | | Leishmania major dihydroorotate dehydrogenase in complex with 5-amino-2-(1H-pyrrol-1-yl)benzonitrile | | Descriptor: | 5-azanyl-2-pyrrol-1-yl-benzenecarbonitrile, Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Cardoso, I.A, Hunter, W.N, Nonato, M.C. | | Deposit date: | 2021-05-20 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Leishmania major dihydroorotate dehydrogenase in complex with 5-amino-2-(1H-pyrrol-1-yl)benzonitrile
To Be Published
|
|
6HY1
 
 | | Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N,N'-Bis(3-aminopropyl)-1,4-cyclohexanediamine after catalysis in crystal | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Sprenger, J, Coertzen, D, Persson, L, Carey, J, Birkholtz, L.M, Louw, B.I. | | Deposit date: | 2018-10-19 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N,N'-Bis(3-aminopropyl)-1,4-cyclohexanediamine after catalysis in crystal
To Be Published
|
|
5X8I
 
 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
5MZL
 
 | |
9M0I
 
 | | Structure of the intermediate of lactoperoxidase formed with thiocynate and hydrogen peroxidase at 1.99 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Viswanathan, V, Maurya, A, Sirohi, H.V, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2025-02-24 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Structure of the intermediate of lactoperoxidase formed with thiocynate and hydrogen peroxidase at 1.99 A resolution.
To Be Published
|
|
3FN4
 
 | | Apo-form of NAD-dependent formate dehydrogenase from bacterium Moraxella sp.C-1 in closed conformation | | Descriptor: | GLYCEROL, NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Polyakov, K.M, Filippova, E.V, Dorovatovskiy, P.V, Tikhonova, T.V, Sadykhov, E.G, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft
Acta Crystallogr.,Sect.D, 65, 2009
|
|
