4GCR
 
 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2X51
 
 | | M6 delta Insert1 | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Squires, G, Houdusse, A. | | Deposit date: | 2010-02-04 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Insert-1 of Myosin Vi in Modulating Nucleotide Affinity.
J.Biol.Chem., 286, 2011
|
|
4PZ3
 
 | |
3G56
 
 | | Structure of the macrolide biosensor protein, MphR(A) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Regulator of macrolide 2'-phosphotransferase I | | Authors: | Zheng, J, Sagar, V, Smolinsky, A, Bourke, C, LaRonde-LeBlanc, N, Cropp, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the macrolide biosensor protein, MphR(A), with and without erythromycin
J.Mol.Biol., 387, 2009
|
|
4KEZ
 
 | | Crystal structure of SsoPox W263F | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
3FOF
 
 | | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*AP*CP*CP*AP*AP*GP*GP*TP*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Laponogov, I, Sohi, M.K, Veselkov, D.A, Pan, X.-S, Sawhney, R, Thompson, A.W, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2008-12-30 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases
Nat.Struct.Mol.Biol., 16, 2009
|
|
7KD7
 
 | |
2X45
 
 | | Crystal Structure of Arg r 1 in complex with histamine | | Descriptor: | ALLERGEN ARG R 1, HISTAMINE | | Authors: | Paesen, G.C, Siebold, C, Syme, N, Harlos, K, Graham, S.C, Hilger, C, Homans, S.W, Hentges, F, Stuart, D.I. | | Deposit date: | 2010-01-28 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Allergen Arg R 1, a Histamine-Binding Lipocalin from a Soft Tick
To be Published
|
|
4KHT
 
 | | Triple helix bundle of GP41 complexed with fab 8066 | | Descriptor: | 8066 heavy chain, 8066 light chain, Gp41 helix | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2013-05-01 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Complexes of neutralizing and non-neutralizing affinity matured Fabs with a mimetic of the internal trimeric coiled-coil of HIV-1 gp41.
Plos One, 8, 2013
|
|
5SMB
 
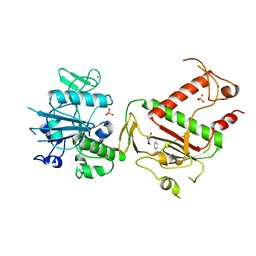 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z419995480 | | Descriptor: | 1-(morpholin-4-yl)-4-phenylbutan-1-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Z0A
 
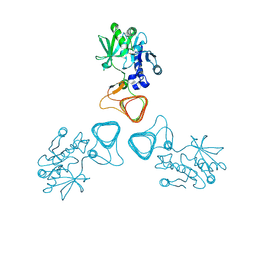 | | ST0452(Y97N)-GlcNAc binding form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual sugar-1-phosphate nucleotidylyltransferase | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
4Q6E
 
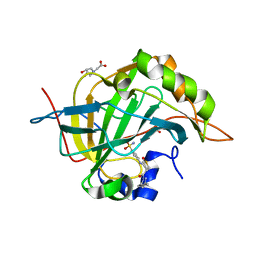 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[3-(3,5-Dimethyl-1H-pyrazol-1-yl)-3-oxopropyl]amino}benzene-1-sulfonamide | | Descriptor: | 4-{[3-(3,5-dimethyl-1H-pyrazol-1-yl)-3-oxopropyl]amino}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 4-Amino-substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Molecules, 19, 2014
|
|
5A8Q
 
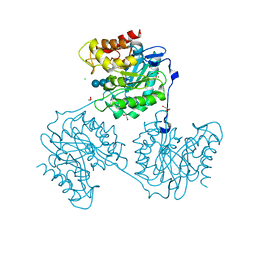 | | Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A obtained by soaking | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
1JB3
 
 | |
3ZIA
 
 | | The structure of F1-ATPase from Saccharomyces cerevisiae inhibited by its regulatory protein IF1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Robinson, G.C, Bason, J.V, Montgomery, M.G, Fearnley, I.M, Mueller, D.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2013-01-07 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of F1-ATPase from Saccharomyces Cerevisiae Inhibited by its Regulatory Protein If1.
Open Biol., 3, 2013
|
|
2QGV
 
 | | Crystal structure of hydrogenase-1 operon protein hyaE from Shigella flexneri. Northeast Structural Genomics Consortium Target SfR170 | | Descriptor: | Hydrogenase-1 operon protein hyaE | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Benach, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hydrogenase-1 operon protein hyaE from Shigella flexneri.
To be Published
|
|
5SLU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1796014543 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4E28
 
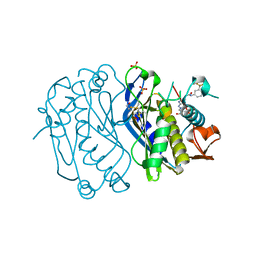 | | Structure of human thymidylate synthase in inactive conformation with a novel non-peptidic inhibitor | | Descriptor: | 2-{(2Z,5S)-4-hydroxy-2-[(2E)-(2-hydroxybenzylidene)hydrazinylidene]-2,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, 2-{(5S)-2-[(2E)-2-(2-hydroxybenzylidene)hydrazinyl]-4-oxo-4,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, SULFATE ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-03-07 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Inhibitor of ovarian cancer cells growth by virtual screening: a new thiazole derivative targeting human thymidylate synthase.
J.Med.Chem., 55, 2012
|
|
1X81
 
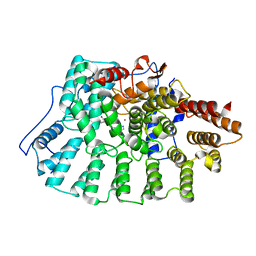 | | Farnesyl transferase structure of Jansen compound | | Descriptor: | 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, Protein farnesyltransferase beta subunit, Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit, ... | | Authors: | Li, Q, Claiborne, A, Li, T, Hasvold, L, Stoll, V.S, Muchmore, S, Jakob, C.G, Gu, W, Cohen, J, Hutchins, C, Frost, D, Rosenberg, S.H, Sham, H.L. | | Deposit date: | 2004-08-16 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, synthesis, and activity of 4-quinolone and pyridone compounds as nonthiol-containing farnesyltransferase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
7JNX
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)pivalamide | | Descriptor: | 2,2-dimethyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.286 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JO0
 
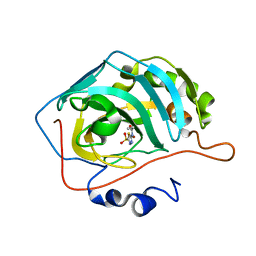 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propionamide | | Descriptor: | Carbonic anhydrase 2, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, ZINC ION | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
1C9Y
 
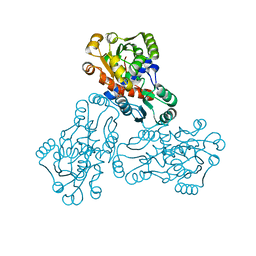 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CATALYTIC MECHANISM | | Descriptor: | NORVALINE, ORNITHINE CARBAMOYLTRANSFERASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Yu, X, Morizono, H, Tuchman, M, Allewell, N.M. | | Deposit date: | 1999-08-03 | | Release date: | 2000-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
Proteins, 39, 2000
|
|
7JJH
 
 | |
4NR5
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Pike, A.W, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
6MLH
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with GLUCOPYRANOSYL-1-METHYL-PHOSPHONIC ACID | | Descriptor: | (1S)-1,5-anhydro-1-(phosphonomethyl)-D-glucitol, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-09-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
