2QVU
 
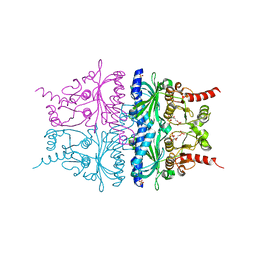 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Mg2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
2QVR
 
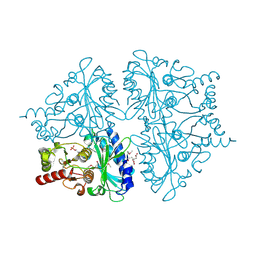 | | E. coli Fructose-1,6-bisphosphatase: Citrate, Fru-2,6-P2, and Mg2+ bound | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, CITRIC ACID, Fructose-1,6-bisphosphatase, ... | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of Mammalian and Bacterial Fructose-1,6-bisphosphatase Reveal the Basis for Synergism in AMP/Fructose 2,6-Bisphosphate Inhibition.
J.Biol.Chem., 282, 2007
|
|
9ELX
 
 | |
4CVM
 
 | |
6CVT
 
 | | Human Aprataxin (Aptx) V263G bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVP
 
 | | Human Aprataxin (Aptx) R199H bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
3RK0
 
 | | X-ray crystal Structure of the putative N-type ATP pyrophosphatase (PF0828) in complex with AMP from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR23 | | Descriptor: | ADENOSINE MONOPHOSPHATE, N-type ATP pyrophosphatase superfamily | | Authors: | Forouhar, F, Saadat, N, Hussain, M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Ma, L, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-16 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A large conformational change in the putative ATP pyrophosphatase PF0828 induced by ATP binding.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5E7J
 
 | |
5YFU
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Pyrococcus horikoshii OT3 in complex with ribulose-1,5-bisphosphate and AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, POTASSIUM ION, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
5YG8
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Pyrococcus horikoshii OT3 in complex with ribulose-1,5-bisphosphate, AMP and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
2QRK
 
 | | Crystal Structure of AMP-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Saccharopine dehydrogenase [NAD+, L-lysine-forming | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
4WF2
 
 | | Structure of E. coli BirA G142A bound to biotinol-5'-AMP | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Bifunctional ligase/repressor BirA | | Authors: | Eginton, C, Beckett, D, Wade, H. | | Deposit date: | 2014-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Allosteric Coupling via Distant Disorder-to-Order Transitions.
J.Mol.Biol., 427, 2015
|
|
4CVL
 
 | | PaMurF in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Ruane, K.M, Majce, V. | | Deposit date: | 2014-03-28 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Pamurf in Complex with AMP-Pnp
To be Published
|
|
4YP1
 
 | | Misting with CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Chin, K.H, Chen, C.K, Tu, Z.I, Chou, S.H. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into the Distinct Binding Mode of Cyclic Di-AMP with SaCpaA_RCK.
Biochemistry, 54, 2015
|
|
4TY0
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with linear intermediate 5' pppA(3',5')pG | | Descriptor: | ACETATE ION, Cyclic AMP-GMP synthase, MAGNESIUM ION, ... | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4HPT
 
 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying complete phosphoryl transfer of AMP-PNP onto a substrate peptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | Deposit date: | 2012-10-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
4TXZ
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with nonhydrolyzable GTP | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4S1C
 
 | |
5YFV
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant C135S from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
5YFW
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and AMP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
5YGA
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate, AMP and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
5YFX
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and AMP | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, Ribose 1,5-bisphosphate isomerase | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
5YG9
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant C135S from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate, AMP and GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
4UN0
 
 | | Crystal structure of the human CDK12-cyclinK complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Chaikuad, A, Goubin, S, Krojer, T, Sorrell, F.J, Nowak, R, Williams, E, Kopec, J, Mahajan, R.P, Burgess-Brown, N, Carpenter, E.P, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
5F29
 
 | | Structure of RCK domain with cda | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Na+/H+ antiporter-like protein | | Authors: | Chin, K.H. | | Deposit date: | 2015-12-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural Insights into the Distinct Binding Mode of Cyclic Di-AMP with SaCpaA_RCK.
Biochemistry, 54, 2015
|
|
