5WOW
 
 | |
5XG5
 
 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
6EGP
 
 | |
7NBI
 
 | |
4MTP
 
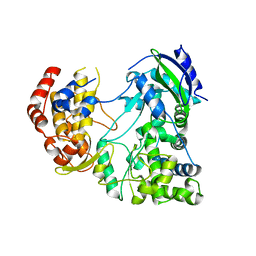 | | RdRp from Japanesese Encephalitis Virus | | Descriptor: | RNA dependent RNA polymerase, ZINC ION | | Authors: | Surana, P, Nair, D.T. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
2P6J
 
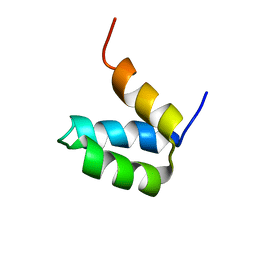 | | Full-sequence computational design and solution structure of a thermostable protein variant | | Descriptor: | designed engrailed homeodomain variant UVF | | Authors: | Shah, P.S, Hom, G.K, Ross, S.A, Lassila, J.K, Crowhurst, K.A, Mayo, S.L. | | Deposit date: | 2007-03-18 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Full-sequence Computational Design and Solution Structure of a Thermostable Protein Variant.
J.Mol.Biol., 372, 2007
|
|
2P05
 
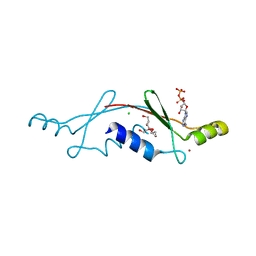 | | Structural Insights into the Evolution of a Non-Biological Protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-02-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
4W5P
 
 | | Prp peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4W67
 
 | | Crystal structure of Prp peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-20 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4V3Q
 
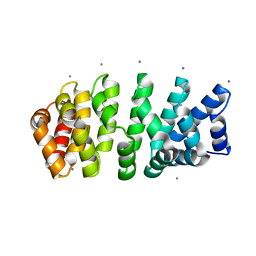 | | Designed armadillo repeat protein with 4 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | CALCIUM ION, GLYCEROL, YIII_M4_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4W71
 
 | | Crystal structure of a prion peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-21 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4V8V
 
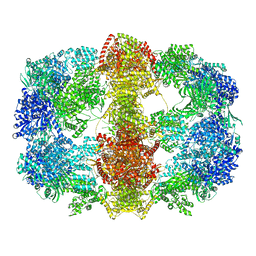 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
4V8W
 
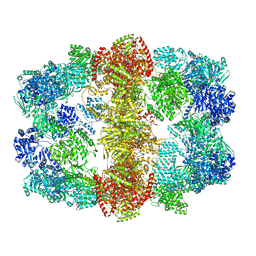 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17.5 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
8Q52
 
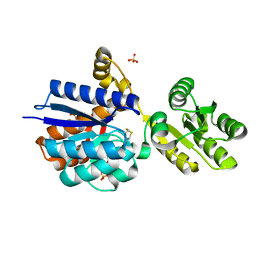 | | A PBP-like protein built from fragments of different folds | | Descriptor: | Leucine-specific-binding protein,Chemotaxis protein CheY, SULFATE ION | | Authors: | Shanmugaratnam, S, Toledo-Patino, S, Goetz, S.K, Farias-Rico, J.A, Hocker, B. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular handcraft of a well-folded protein chimera.
Febs Lett., 598, 2024
|
|
7RX5
 
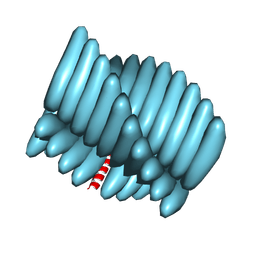 | | Cryo-EM reconstruction of Form1-N2 nanotube (Form I like) | | Descriptor: | F1-N2 nanotube | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
6OV9
 
 | |
6OVS
 
 | | Coiled-coil Trimer with Glu:Tyr:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:Tyr:Lys Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-05-08 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
6OSD
 
 | |
6OLO
 
 | |
2DX4
 
 | |
7CCN
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | Descriptor: | LBT3, LUTETIUM (III) ION | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
5I1N
 
 | | Villin headpiece subdomain with a Gln26 to beta-3-homoglutamine substitution | | Descriptor: | D-Villin headpiece subdomain, Villin-1 | | Authors: | Kreitler, D.F, Mortenson, D.E, Gellman, S.H, Forest, K.T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effects of Single alpha-to-beta Residue Replacements on Structure and Stability in a Small Protein: Insights from Quasiracemic Crystallization.
J.Am.Chem.Soc., 138, 2016
|
|
5I1Z
 
 | | Structure of nvPizza2-H16S58 | | Descriptor: | SULFATE ION, nvPizza2-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broken Symmetry: Partial domain swapping in an artificial trimeric protein
To Be Published
|
|
5IF6
 
 | |
6B17
 
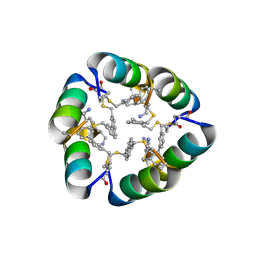 | | Design of a short thermally stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-09-17 | | Release date: | 2018-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
