2VI3
 
 | |
2VI2
 
 | |
2WBZ
 
 | |
2VU6
 
 | |
2VU7
 
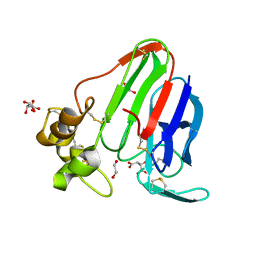 | | Atomic resolution (1.08 A) structure of purified thaumatin I grown in sodium meso-tartrate at 4 C | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, Thaumatin-1 | | Authors: | Jakoncic, J, Asherie, N, Ginsberg, C. | | Deposit date: | 2008-05-21 | | Release date: | 2009-07-14 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Tartrate Chirality Determines Thaumatin Crystal Habit
Cryst.Growth Des., 9, 2009
|
|
2VHK
 
 | |
2VI4
 
 | |
2M5X
 
 | | Novel method of protein purification for structural research. Example of ultra high resolution structure of SPI-2 inhibitor by X-ray and NMR spectroscopy. | | Descriptor: | Silk protease inhibitor 2 | | Authors: | Lenarcic Zivkovic, M, Dvornyk, A, Kludkiewicz, B, Kopera, E, Zagorski-Ostoja, W, Grzelak, K, Zhukov, I, Bal, W. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
2NDH
 
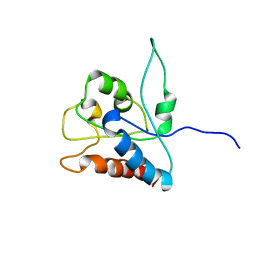 | | NMR solution structure of MAL/TIRAP TIR domain (C116A) | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Lavrencic, P, Mobli, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2NCG
 
 | |
2OZE
 
 | | The Crystal structure of Delta protein of pSM19035 from Streptoccocus pyogenes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Orf delta', ... | | Authors: | Cicek, A, Weihofen, W, Saenger, W. | | Deposit date: | 2007-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation.
Nucleic Acids Res., 36, 2008
|
|
2LT2
 
 | |
2IBS
 
 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position | | Descriptor: | 5'-D(*GP*AP*CP*AP*TP*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Pljevaljcic, G, Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
7JQL
 
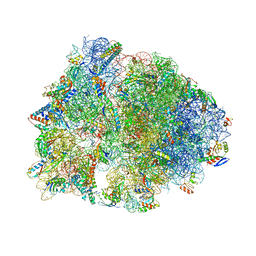 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-001, mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
2M41
 
 | |
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
1SB1
 
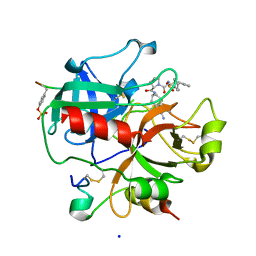 | | Novel Non-Covalent Thrombin Inhibitors Incorporating P1 4,5,6,7-Tetrahydrobenzothiazole Arginine Side Chain Mimetics | | Descriptor: | N-(BENZYLSULFONYL)-3-CYCLOHEXYLALANYL-N-(2-AMINO-1,3-BENZOTHIAZOL-6-YL)PROLINAMIDE, Prothrombin, SODIUM ION, ... | | Authors: | Marinko, P, Krbavcic, A, Mlinsek, G, Solmajer, T, Trampus-Bakija, A, Stegnar, M, Stojan, J, Kikelj, D. | | Deposit date: | 2004-02-09 | | Release date: | 2004-06-08 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel non-covalent thrombin inhibitors incorporating P(1) 4,5,6,7-tetrahydrobenzothiazole arginine side chain mimetics
Eur.J.Med.Chem., 39, 2004
|
|
7B7V
 
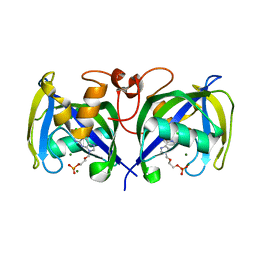 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
7JKZ
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with YF3-126 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKY
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-126 | | Descriptor: | Bromodomain-containing protein 4, N-(1-[1,1-di(pyridin-2-yl)ethyl]-6-{1-methyl-6-oxo-5-[(piperidin-4-yl)amino]-1,6-dihydropyridin-3-yl}-1H-indol-4-yl)ethanesulfonamide, SODIUM ION | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
7JKW
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with ZN1-99 | | Descriptor: | Bromodomain-containing protein 4, N-(6-{5-[(azetidin-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridin-3-yl}-1-[1,1-di(pyridin-2-yl)ethyl]-1H-indol-4-yl)ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
4WT3
 
 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT4
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT5
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form II | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6P05
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with compound 27 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Zhao, J, Gutgesell, L.M, Shen, Z, Dye, K, Dubrovyskyii, O, Zhao, H, Huang, F, Tonetti, D.A, Thatcher, G.R. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Novel Pyrrolopyridone Bromodomain and Extra-Terminal Motif (BET) Inhibitors Effective in Endocrine-Resistant ER+ Breast Cancer with Acquired Resistance to Fulvestrant and Palbociclib.
J.Med.Chem., 63, 2020
|
|
