5JXJ
 
 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3H03
 
 | |
7GU0
 
 | |
7A6A
 
 | | 1.15 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.15 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
7D56
 
 | |
7GU1
 
 | |
2YCK
 
 | | methyltransferase bound with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 5-METHYLTETRAHYDROFOLATE CORRINOID/IRON SULFUR PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Goetzl, S, Jeoung, J.-H, Hennig, S.E, Dobbek, H. | | Deposit date: | 2011-03-16 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Electron and Methyl-Group Transfer in a Methyltransferase System Operating in the Reductive Acetyl-Coa Pathway
J.Mol.Biol., 411, 2011
|
|
7OSH
 
 | | ABC Transporter complex NosDFYL, R-domain 2 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
2L8Y
 
 | | Solution structure of the E. coli outer membrane protein RcsF (periplasmatic domain) | | Descriptor: | Protein rcsF | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Lohr, F, Doetsch, V. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF.
J.Biol.Chem., 286, 2011
|
|
7D5V
 
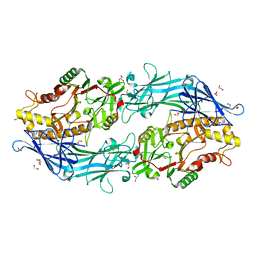 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | Authors: | Akimoto, M, Mashimo, R, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7GT1
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000209a | | Descriptor: | (2S)-2-(2-chloro-6-fluorophenyl)-2,3-dihydroquinazolin-4(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GUB
 
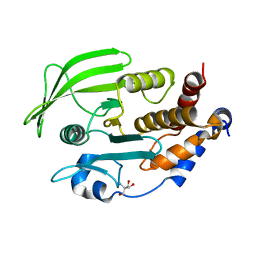 | |
7OSJ
 
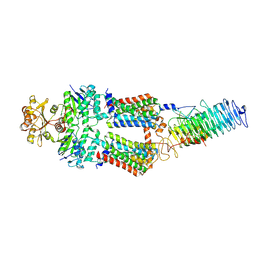 | | ABC Transporter complex NosDFYL, membrane anchor | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
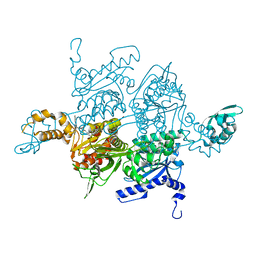
3H0Q
 
 | |
7GUA
 
 | |
7DAN
 
 | |
2LAF
 
 | |
7GT6
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000530a | | Descriptor: | (3aS,8aS)-6-benzoyloctahydropyrrolo[3,4-d]azepin-1(2H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
2YZE
 
 | | Crystal structure of uricase from Arthrobacter globiformis | | Descriptor: | (dihydroxyboranyloxy-hydroxy-boranyl)oxylithium, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
7GUC
 
 | |
7D4Y
 
 | |
7OS7
 
 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant
To Be Published
|
|
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mashimo, R, Akimoto, M, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7GTE
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000646b | | Descriptor: | (5S)-5-(trifluoromethyl)-1,4-diazepane, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
3GUR
 
 | | Crystal Structure of mu class glutathione S-transferase (GSTM2-2) in complex with glutathione and 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase Mu 2, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
