1ZDX
 
 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZDY
 
 | |
1ZE1
 
 | |
1ZE2
 
 | |
1ZE3
 
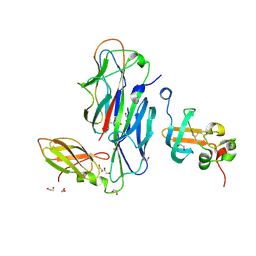 | | Crystal Structure of the Ternary Complex of FIMD (N-Terminal Domain) with FIMC and the Pilin Domain of FIMH | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein fimC, FimH protein, ... | | Authors: | Nishiyama, M, Horst, R, Eidam, O, Herrmann, T, Ignatov, O, Vetsch, M, Bettendorff, P, Jelesarov, I, Grutter, M.G, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZE7
 
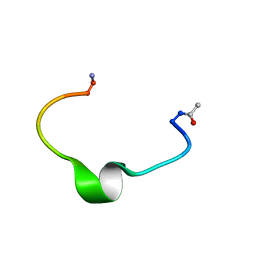 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide in water solution at pH 6.5 | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2013-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
1ZE8
 
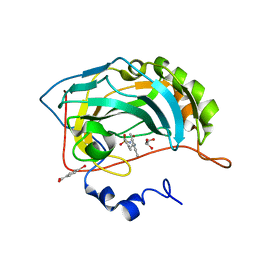 | | Carbonic anhydrase II in complex with a membrane-impermeant sulfonamide inhibitor | | Descriptor: | 1-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}-2,4,6-TRIMETHYLPYRIDINIUM, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase II, ... | | Authors: | Menchise, V, De Simone, G, Alterio, V, Di Fiore, A, Pedone, C, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2005-04-18 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbonic anhydrase inhibitors: stacking with Phe131 determines active site binding region of inhibitors as exemplified by the X-ray crystal structure of a membrane-impermeant antitumor sulfonamide complexed with isozyme II
J.Med.Chem., 48, 2005
|
|
1ZE9
 
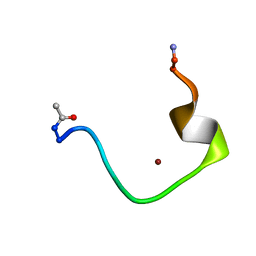 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide complexed with a zinc (II) cation | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein, ZINC ION | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2013-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
1ZEA
 
 | | Structure of the anti-cholera toxin antibody Fab fragment TE33 in complex with a D-peptide | | Descriptor: | CITRIC ACID, monoclonal anti-cholera toxin IGG1 KAPPA antibody, H chain, ... | | Authors: | Scheerer, P, Krauss, N, Wessner, H, Scholz, C, Otte, L, Seifert, M, Kramer, A, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2005-04-18 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an anti-cholera toxin antibody Fab in complex with an epitope-derived D-peptide: a case of polyspecific recognition.
J.Mol.Recognit., 20, 2007
|
|
1ZEB
 
 | | X-ray structure of alkaline phosphatase from human placenta in complex with 5'-AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1ZEC
 
 | | NMR Solution structure of NEF1-25, 20 structures | | Descriptor: | NEF1-25 | | Authors: | Barnham, K.J, Monks, S.A, Hinds, M.G, Azad, A.A, Norton, R.S. | | Deposit date: | 1996-12-18 | | Release date: | 1998-01-07 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Biochemistry, 36, 1997
|
|
1ZED
 
 | | Alkaline phosphatase from human placenta in complex with p-nitrophenyl-phosphonate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1ZEE
 
 | | X-Ray Crystal Structure of Protein SO4414 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR52. | | Descriptor: | hypothetical protein SO4414 | | Authors: | Forouhar, F, Abashidze, M, Vorobiev, S.M, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of the Hypothetical Protein SO4414 from Shewanella oneidensis, NESG Target SoR52
To be Published
|
|
1ZEF
 
 | | structure of alkaline phosphatase from human placenta in complex with its uncompetitive inhibitor L-Phe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEH
 
 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEI
 
 | | CROSS-LINKED B28 ASP INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEJ
 
 | | Crystal structure of the 3-hydroxyacyl-coa dehydrogenase (hbd-9, af2017) from archaeoglobus fulgidus dsm 4304 at 2.00 A resolution | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3-hydroxyacyl-CoA dehydrogenase, CHLORIDE ION | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-hydroxyacyl-CoA dehydrogenase (HBD-9) (np_070841.1) from Archaeoglobus fulgidus at 2.00 A resolution
To be published
|
|
1ZEL
 
 | | Crystal structure of RV2827C protein from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Janowski, R, Panjikar, S, Mueller-dieckmann, J, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-04-19 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural analysis reveals DNA binding properties of Rv2827c, a hypothetical protein from Mycobacterium tuberculosis.
J Struct Funct Genomics, 10, 2009
|
|
1ZEM
 
 | |
1ZEN
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Descriptor: | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE, ZINC ION | | Authors: | Cooper, S.J, Leonard, G.A, Hunter, W.N. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold.
Structure, 4, 1996
|
|
1ZEO
 
 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
1ZEQ
 
 | | 1.5 A Structure of apo-CusF residues 6-88 from Escherichia coli | | Descriptor: | Cation efflux system protein cusF | | Authors: | Loftin, I.R, Franke, S, Roberts, S.A, Weichsel, A, Heroux, A, Montfort, W.R, Rensing, C, McEvoy, M.M. | | Deposit date: | 2005-04-19 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Copper-Binding Fold for the Periplasmic Copper Resistance Protein CusF.
Biochemistry, 44, 2005
|
|
1ZES
 
 | | BeF3- activated PhoB receiver domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Bachhawat, P, Montelione, G.T, Stock, A.M. | | Deposit date: | 2005-04-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Activation for Transcription Factor PhoB Suggested by Different Modes of Dimerization in the Inactive and Active States.
Structure, 13, 2005
|
|
1ZET
 
 | |
