1YUS
 
 | | Solution structure of apo-S100A13 | | Descriptor: | S100 calcium binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUT
 
 | | Solution structure of Calcium-S100A13 (minimized mean structure) | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUU
 
 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUW
 
 | |
1YUX
 
 | | Mixed valant state of nigerythrin | | Descriptor: | FE (II) ION, FE (III) ION, Nigerythrin | | Authors: | Iyer, R.B, Silaghi-Dumitrescu, R, Kurtz, D.M, Lanzilotta, W.N. | | Deposit date: | 2005-02-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins.
J.Biol.Inorg.Chem., 10, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YUZ
 
 | | Partially Reduced State of Nigerythrin | | Descriptor: | FE (II) ION, FE (III) ION, Nigerythrin | | Authors: | Iyer, R.B, Silaghi-Dumitrescu, R, Kurtz, D.M, Lanzilotta, W.N. | | Deposit date: | 2005-02-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins.
J.Biol.Inorg.Chem., 10, 2005
|
|
1YV0
 
 | | Crystal structure of skeletal muscle troponin in the Ca2+-free state | | Descriptor: | MAGNESIUM ION, Troponin C, skeletal muscle, ... | | Authors: | Vinogradova, M.V, Stone, D.B, Malanina, G.G, Karatzaferi, C, Cooke, R, Mendelson, R.A, Fletterick, R.J. | | Deposit date: | 2005-02-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Ca2+-regulated structural changes in troponin
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1YV1
 
 | | Fully reduced state of nigerythrin (all ferrous) | | Descriptor: | FE (II) ION, Nigerythrin | | Authors: | Iyer, R.B, Silaghi-Dumitrescu, R, Kurtz, D.M, Lanzilotta, W.N. | | Deposit date: | 2005-02-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins.
J.Biol.Inorg.Chem., 10, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YV3
 
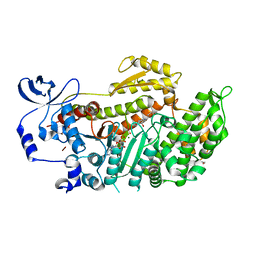 | | The structural basis of blebbistatin inhibition and specificity for myosin II | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Smith, R, Rayment, I. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of blebbistatin inhibition and specificity for myosin II.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YV4
 
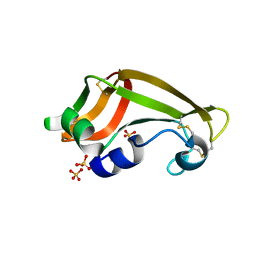 | | X-ray structure of M23L onconase at 100K | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV5
 
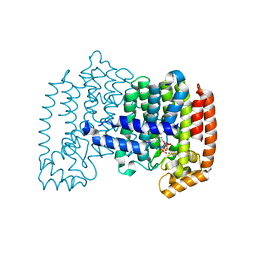 | | Human farnesyl diphosphate synthase complexed with Mg and risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION, ... | | Authors: | Kavanagh, K.L, Guo, K, Von Delft, F, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1YV6
 
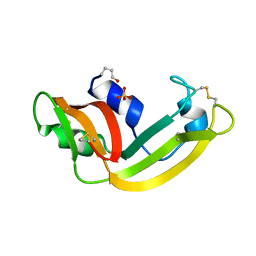 | | X-ray structure of M23L onconase at 298K | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV7
 
 | | X-ray structure of (C87S,des103-104) onconase | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV8
 
 | |
1YV9
 
 | |
1YVA
 
 | | NMR solution structure of crambin in DPC micelles | | Descriptor: | Crambin | | Authors: | Ahn, H.-C, Markley, J.L. | | Deposit date: | 2005-02-15 | | Release date: | 2006-03-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Water-Insoluble Protein Crambin in Dodecylphosphocholine Micelles and Its Minimal Solvent-Exposed Surface
J.Am.Chem.Soc., 128, 2006
|
|
1YVB
 
 | | the Plasmodium falciparum Cysteine Protease Falcipain-2 | | Descriptor: | Cystatin, GLYCEROL, falcipain 2 | | Authors: | Wang, S.X. | | Deposit date: | 2005-02-15 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for unique mechanisms of folding and hemoglobin binding by a malarial protease.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1YVC
 
 | | Solution structure of the conserved protein from the gene locus MMP0076 of Methanococcus maripaludis. Northeast Structural Genomics target MrR5. | | Descriptor: | MrR5 | | Authors: | Northeast Structural Genomics Consortium (NESG), Rossi, P, Aramini, J.M, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: |
To be Published
|
|
1YVD
 
 | | GppNHp-Bound Rab22 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-22A | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-15 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YVE
 
 | | ACETOHYDROXY ACID ISOMEROREDUCTASE COMPLEXED WITH NADPH, MAGNESIUM AND INHIBITOR IPOHA (N-HYDROXY-N-ISOPROPYLOXAMATE) | | Descriptor: | ACETOHYDROXY ACID ISOMEROREDUCTASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Biou, V, Dumas, R, Cohen-Addad, C, Douce, R, Job, D, Pebay-Peyroula, E. | | Deposit date: | 1996-10-11 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of plant acetohydroxy acid isomeroreductase complexed with NADPH, two magnesium ions and a herbicidal transition state analog determined at 1.65 A resolution.
EMBO J., 16, 1997
|
|
1YVF
 
 | | Hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00729145 | | Descriptor: | (2Z)-2-(BENZOYLAMINO)-3-[4-(2-BROMOPHENOXY)PHENYL]-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M.L, Nugent, R.A, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R.A, Kilkuskie, R.E, Kopta, L.A, Schwende, F.J. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 1: Evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YVG
 
 | | Structural analysis of the catalytic domain of tetanus neurotoxin | | Descriptor: | Tetanus toxin, light chain, ZINC ION | | Authors: | Rao, K.N, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the catalytic domain of tetanus neurotoxin.
Toxicon, 45, 2005
|
|
1YVH
 
 | |
