5VD0
 
 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH MK1775 | | Descriptor: | 1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, DI(HYDROXYETHYL)ETHER, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
3L5D
 
 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3KOQ
 
 | |
6N7C
 
 | | Structure of the human JAK1 kinase domain with compound 56 | | Descriptor: | GLYCEROL, N-[5-(3-methoxynaphthalen-2-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5FE2
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment BR013 (fragment 3) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3~{H}-isoindol-1-one, Histone acetyltransferase KAT2B | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE9
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1122 (compound 13) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-(1,4-dimethyl-2-oxidanylidene-quinolin-7-yl)methanesulfonamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
6DA3
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2R)-1-oxo-3-phenyl-1-{[(pyridin-3-yl)methyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
5VIC
 
 | | Crystal structure of anti-Zika antibody Z004 bound to DENV-1 Envelope protein DIII | | Descriptor: | Dengue 1 Envelope DIII domain, Fab heavy chain, Fab light chain | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
1OJH
 
 | | Crystal structure of NblA from PCC 7120 | | Descriptor: | 1,2-ETHANEDIOL, NBLA | | Authors: | Bienert, R, Baier, K, Lockau, W, Heinemann, U. | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Nbla from Anabaena Sp. Pcc 7120, a Small Protein Playing a Key Role in Phycobilisome Degradation.
J.Biol.Chem., 281, 2006
|
|
1XRW
 
 | | Solution Structure of a Platinum-Acridine Modified Octamer | | Descriptor: | 1-[2-(ACRIDIN-9-YLAMINO)ETHYL]-1,3-DIMETHYLTHIOUREA-PLATINUM(II)-ETHANE-1,2-DIAMINE, 5'-D(*CP*CP*TP*CP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Baruah, H, Wright, M.W, Bierbach, U. | | Deposit date: | 2004-10-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Study of a DNA Duplex Containing the Guanine-N7 Adduct Formed by a Cytotoxic Platinum-Acridine Hybrid Agent
Biochemistry, 44, 2005
|
|
1Z9E
 
 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | Descriptor: | PC4 and SFRS1 interacting protein 2 | | Authors: | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | Deposit date: | 2005-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
6K9W
 
 | |
5FDC
 
 | | Crystal structure of Human Carbonic Anhydrase II in complex with the anticonvulsant sulfamide JNJ-26990990 and its S,S-dioxide analog. | | Descriptor: | 3-[(sulfamoylamino)methyl]-1-benzothiophene, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Di Fiore, A, De Simone, G, Alterio, V, Riccio, V, Winum, J.-Y, Carta, F, Supuran, C.T. | | Deposit date: | 2015-12-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The anticonvulsant sulfamide JNJ-26990990 and its S,S-dioxide analog strongly inhibit carbonic anhydrases: solution and X-ray crystallographic studies.
Org.Biomol.Chem., 14, 2016
|
|
2QF4
 
 | |
3KOG
 
 | |
5WE8
 
 | |
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
5BWB
 
 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA IN COMPLEX WITH THE BIS-IMIDAZOLIUM OXIME 2BIM-7 | | Descriptor: | (Z,Z)-[heptane-1,7-diylbis(1H-imidazol-1-yl-2-ylidene)]bis(N-hydroxymethanamine), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Legler, P.M, Millard, C.B. | | Deposit date: | 2015-06-07 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A conformational change in the peripheral anionic site of Torpedo californica acetylcholinesterase induced by a bis-imidazolium oxime.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7GNS
 
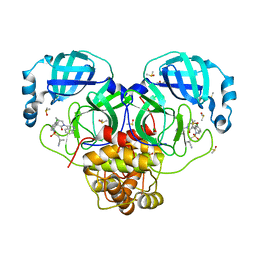 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-50a80394-1 (Mpro-P3050) | | Descriptor: | 1-{[(3'S,4'R)-6-chloro-1'-(isoquinolin-4-yl)-4'-methyl-2'-oxo-1H-spiro[isoquinoline-4,3'-pyrrolidine]-2(3H)-sulfonyl]methyl}cyclopropane-1-carbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4DFE
 
 | |
5CF8
 
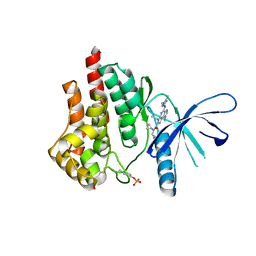 | | CRYSTAL STRUCTURE OF JANUS KINASE 2 IN COMPLEX WITH N,N-DICYCLOPROPYL-10-ETHYL-7-[(3-METHOXYPROPYL)AMINO] -3-METHYL-3,5,8,10-TETRAAZATRICYCLO[7.3.0.0,6] DODECA-1(9),2(6),4,7,11-PENTAENE-11-CARBOXAMIDE | | Descriptor: | N,N-dicyclopropyl-4-[(1,5-dimethyl-1H-pyrazol-3-yl)amino]-6-ethyl-1-methyl-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Sack, J.S. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Highly Selective JAK2 Inhibitor, BMS-911543, for the Treatment of Myeloproliferative Neoplasms.
Acs Med.Chem.Lett., 6, 2015
|
|
397D
 
 | |
3L1B
 
 | | Complex Structure of FXR Ligand-binding domain with a tetrahydroazepinoindole compound | | Descriptor: | 1-methylethyl 8-fluoro-1,1-dimethyl-3-{[4-(3-morpholin-4-ylpropoxy)phenyl]carbonyl}-1,2,3,6-tetrahydroazepino[4,5-b]indole-5-carboxylate, Farnesoid X receptor | | Authors: | Xu, W, Lundquist, J.T. | | Deposit date: | 2009-12-11 | | Release date: | 2010-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Physiochemical Properties of the Tetrahydroazepinoindole Series of Farnesoid X Receptor (FXR) Agonists: Beneficial Modulation of Lipids in Primates.
J.Med.Chem., 53, 2010
|
|
1X95
 
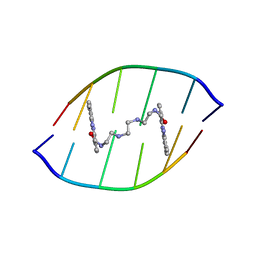 | | Solution structure of the DNA-hexamer ATGCAT complexed with DNA Bis-intercalating Anticancer Drug XR5944 (MLN944) | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, 5'-D(*AP*TP*GP*CP*AP*T)-3' | | Authors: | Dai, J, Punchihewa, C, Mistry, P, Ooi, A.T, Yang, D. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug.
J.Biol.Chem., 279, 2004
|
|
1D78
 
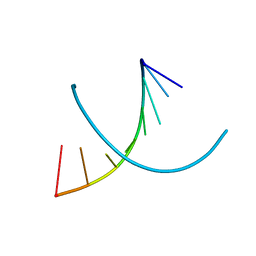 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
