6RF4
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF3
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, SODIUM ION, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
5NA1
 
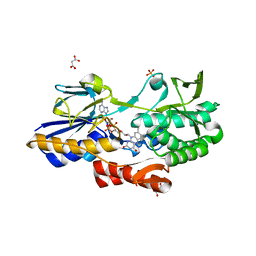 | | NADH:quinone oxidoreductase (NDH-II) from Staphylococcus aureus - holoprotein structure - 2.32 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, NADH dehydrogenase-like protein SAOUHSC_00878, ... | | Authors: | Brito, J.A, Athayde, D, Sousa, F.M, Sena, F.V, Pereira, M.M, Archer, M. | | Deposit date: | 2017-02-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The key role of glutamate 172 in the mechanism of type II NADH:quinone oxidoreductase of Staphylococcus aureus.
Biochim. Biophys. Acta, 1858, 2017
|
|
5NAE
 
 | |
5NB3
 
 | | High resolution C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
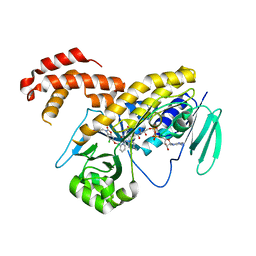
5NGZ
 
 | | Ube2T in complex with fragment EM04 | | Descriptor: | 1-(1,3-benzothiazol-2-yl)methanamine, Ubiquitin-conjugating enzyme E2 T | | Authors: | Morreale, F.E, Bortoluzzi, A, Chaugule, V.K, Arkinson, C, Walden, H, Ciulli, A. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric Targeting of the Fanconi Anemia Ubiquitin-Conjugating Enzyme Ube2T by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5NH5
 
 | |
5NHF
 
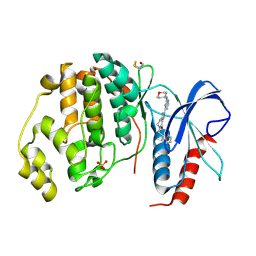 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-5-(phenylmethyl)-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
6RG6
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[3-(6-azanyl-9~{H}-purin-8-yl)prop-2-ynoxymethyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
6RG7
 
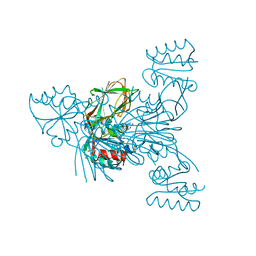 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-(6-azanyl-9~{H}-purin-8-yl)prop-2-ynyl-methyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
5NHJ
 
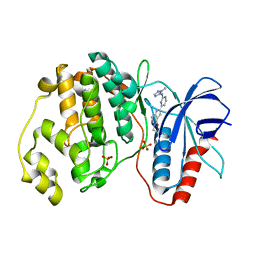 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHP
 
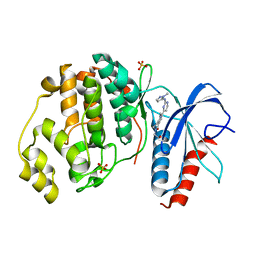 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-1-methyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
6RGA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(aminomethyl)-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
5NHU
 
 | |
6RGB
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynylamino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
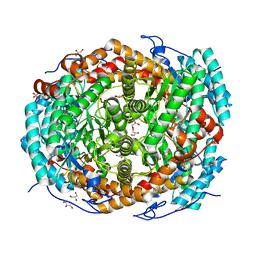
5NGD
 
 | | Crystal structure of the V325G mutant of the bacteriophage G20C portal protein | | Descriptor: | Portal protein | | Authors: | Pascoa, T, Jenkins, H.T, Chechik, M, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Porphyrin-Assisted Docking of a Thermophage Portal Protein into Lipid Bilayers: Nanopore Engineering and Characterization
Acs Nano, 11, 2017
|
|
6RP3
 
 | | Truncated Norcoclaurine synthase with reaction intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-[[(2~{R})-2-phenylpropyl]amino]ethyl]benzene-1,2-diol, S-norcoclaurine synthase | | Authors: | Keep, N.H, Roddan, R, Sula, A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Acceptance and Kinetic Resolution of alpha-Methyl-Substituted Aldehydes by Norcoclaurine Synthases
Acs Catalysis, 2019
|
|
5NGR
 
 | | Crystal structure of human MTH1 in complex with fragment inhibitor 8-(methylsulfanyl)-7H-purin-6-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-methylsulfanyl-7~{H}-purin-6-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
6RGK
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-055 | | Descriptor: | 3-[5-[(4~{a}~{R},8~{a}~{S})-3-cycloheptyl-4-oxidanylidene-4~{a},5,6,7,8,8~{a}-hexahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-butyl-prop-2-ynamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
5NIJ
 
 | |
6RH0
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 5.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
5NH2
 
 | |
6RR6
 
 | | Structure of 100% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RHM
 
 | | Room temperature data of Galectin-3C in complex with a pair of enantiomeric ligands: S enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{S})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Are crystallographic B-factors suitable for calculating protein conformational entropy?
Phys Chem Chem Phys, 21, 2019
|
|
