1BZW
 
 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
4PXJ
 
 | |
8G2O
 
 | | Fab structure - Anti-ApoE-7C11 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, anti-ApoE-7C11 heavy chain, anti-ApoE-7C11 light chain | | Authors: | Marino, C, Arboleda-Velasquez, J.F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | APOE Christchurch-mimetic therapeutic antibody reduces APOE-mediated toxicity and tau phosphorylation.
Alzheimers Dement, 20, 2024
|
|
1U2U
 
 | |
4M9C
 
 | | WeeI from Acinetobacter baumannii AYE | | Descriptor: | Bacterial transferase hexapeptide (Three repeats) family protein | | Authors: | Morrison, M.J, Imperiali, B. | | Deposit date: | 2013-08-14 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical analysis and structure determination of bacterial acetyltransferases responsible for the biosynthesis of UDP-N,N'-diacetylbacillosamine.
J.Biol.Chem., 288, 2013
|
|
3GUS
 
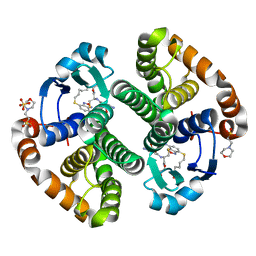 | | Crystal strcture of human Pi class glutathione S-transferase GSTP1-1 in complex with 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
3IE3
 
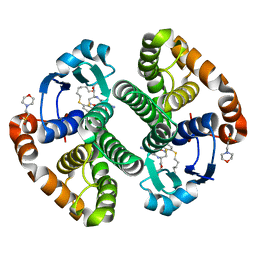 | | Structural basis for the binding of the anti-cancer compound 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) to human glutathione S-transferases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
6UCM
 
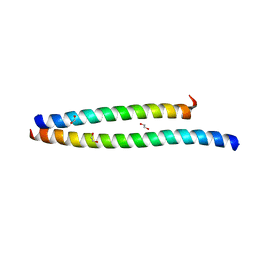 | | Transcription factor DeltaFosB bZIP domain self-assembly, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6UCL
 
 | |
2ZDU
 
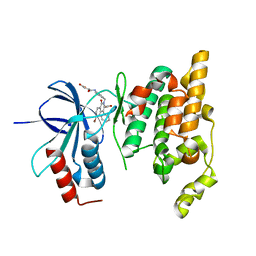 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(4-{[6-bromo-3-(methoxycarbonyl)-1-oxo-4-phenylisoquinolin-2(1H)-yl]methyl}phenyl)amino]-4-oxobutanoic acid, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Ohra, T, Itoh, F, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (1)
Bioorg.Med.Chem., 16, 2008
|
|
6RFP
 
 | | ERK2 MAP kinase with mutations at Helix-G | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Livnah, O, Eitan-Wexler, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop.
J.Biol.Chem., 295, 2020
|
|
2ZV6
 
 | | Crystal structure of human squamous cell carcinoma antigen 1 | | Descriptor: | Serpin B3 | | Authors: | Zheng, B, Matoba, Y, Katagiri, C, Hibino, T, Sugiyama, M. | | Deposit date: | 2008-11-01 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SCCA1 and insight about the interaction with JNK1
Biochem.Biophys.Res.Commun., 380, 2009
|
|
2Y9Q
 
 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | MAP KINASE-INTERACTING SERINE/THREONINE-PROTEIN KINASE 1, MITOGEN-ACTIVATED PROTEIN KINASE 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Barkai, T, Garai, A, Toeroe, I, Remenyi, A. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
3V6S
 
 | |
3V6R
 
 | |
3T7C
 
 | |
3TSC
 
 | |
3PGX
 
 | |
3PXX
 
 | |
3S55
 
 | |
3N1B
 
 | | C-terminal domain of Vps54 subunit of the GARP complex | | Descriptor: | Vacuolar protein sorting-associated protein 54 | | Authors: | Perez-Victoria, F.J, Abascal-Palacios, G, Tascon, I, Kajava, A, Pioro, E.P, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2010-05-15 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TEI
 
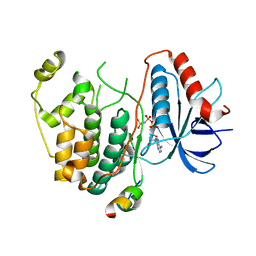 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Remenyi, A. | | Deposit date: | 2011-08-15 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove.
Sci.Signal., 5, 2012
|
|
4FMQ
 
 | |
1TR2
 
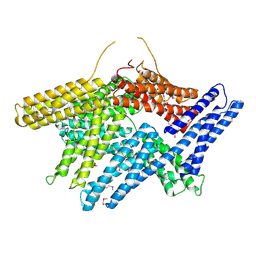 | | Crystal structure of human full-length vinculin (residues 1-1066) | | Descriptor: | VINCULIN ISOFORM 1 | | Authors: | Borgon, R.A, Vonrhein, C, Bricogne, G, Bois, P.R, Izard, T. | | Deposit date: | 2004-06-19 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Vinculin
Structure, 12, 2004
|
|
1NF3
 
 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | Descriptor: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | Authors: | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
