1NSL
 
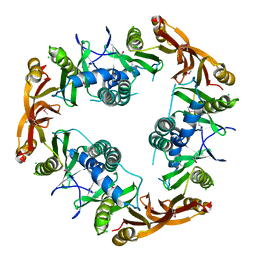 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
3FTQ
 
 | |
3FR0
 
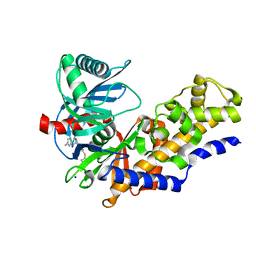 | | Human glucokinase in complex with 2-amino benzamide activator | | Descriptor: | 2-amino-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of novel and potent 2-amino benzamide derivatives as allosteric glucokinase activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1QHJ
 
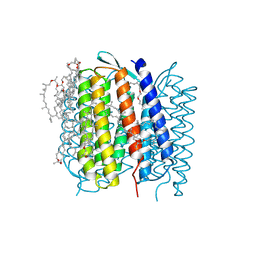 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
3GD6
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate | | Descriptor: | Muconate cycloisomerase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate.
To be Published
|
|
1QKP
 
 | | HIGH RESOLUTION X-RAY STRUCTURE OF AN EARLY INTERMEDIATE IN THE BACTERIORHODOPSIN PHOTOCYCLE | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Edman, K, Nollert, P, Royant, A, Belrhali, H, Pebay-Peyroula, E, Hajdu, J, Neutze, R, Landau, E.M. | | Deposit date: | 1999-07-30 | | Release date: | 1999-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle.
Nature, 401, 1999
|
|
4PV6
 
 | | Crystal Structure Analysis of Ard1 from Thermoplasma volcanium | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-terminal acetyltransferase complex subunit [ARD1] | | Authors: | Ma, C, Lee, S.J, Lee, B.J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A.
Biochim.Biophys.Acta, 1844, 2014
|
|
1R2N
 
 | | NMR structure of the all-trans retinal in dark-adapted Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, terLaak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4NPT
 
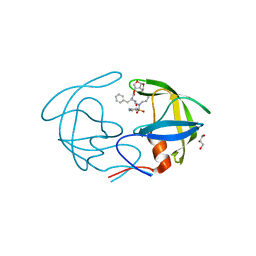 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
4NYH
 
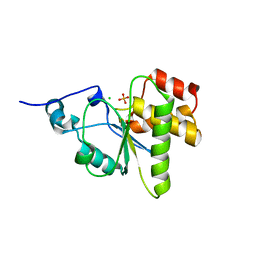 | | Orthorhombic crystal form of pir1 dual specificity phosphatase core | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Sankhala, R.S, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2013-12-10 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Human PIR1, an Atypical Dual-Specificity Phosphatase.
Biochemistry, 53, 2014
|
|
1P3C
 
 | | Glutamyl endopeptidase from Bacillus intermedius | | Descriptor: | glutamyl-endopeptidase | | Authors: | Meijers, R, Blagova, E.V, Levdikov, V.M, Rudenskaya, G.N, Chestukhina, G.G, Akimkina, T.V, Kostrov, S.V, Lamzin, V.S, Kuranova, I.P. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation.
Biochemistry, 43, 2004
|
|
4NXC
 
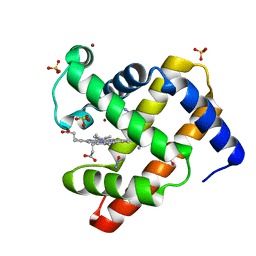 | | SPERM WHALE MYOGLOBIN UNDER 30 BAR NITROUS Oxide | | Descriptor: | Myoglobin, NITROUS OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Colloc'h, N, Prange, T, Vallone, B. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystallographic Studies with Xenon and Nitrous Oxide Provide Evidence for Protein-dependent Processes in the Mechanisms of General Anesthesia
Anesthesiology, 121, 2014
|
|
4PQ8
 
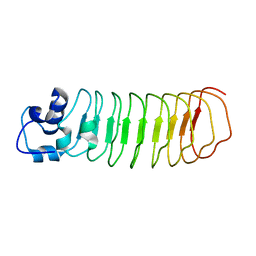 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
1Q9O
 
 | | S45-18 Fab Unliganded | | Descriptor: | MAGNESIUM ION, S45-2 Fab (IgG1k) heavy chain, S45-2 Fab (IgG1k) light chain | | Authors: | Nguyen, H.P, Seto, N.O, MacKenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2003-08-25 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
1QKE
 
 | | ERABUTOXIN | | Descriptor: | ERABUTOXIN A, SULFATE ION | | Authors: | Nastopoulos, V, Kanellopoulos, P.N, Tsernoglou, D. | | Deposit date: | 1998-01-16 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3IGF
 
 | | Crystal Structure of the All4481 protein from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR300 | | Descriptor: | All4481 protein | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target NsR300
To be Published
|
|
3IK1
 
 | | Lactobacillus casei Thymidylate Synthase in Ternary Complex with dUMP and the Phtalimidic Derivative 20C | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-hydroxy-4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid, Thymidylate synthase | | Authors: | Pozzi, C, Cancian, L, Leone, R, Luciani, R, Ferrari, S, Mangani, S, Costi, M.P. | | Deposit date: | 2009-08-05 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
4NXA
 
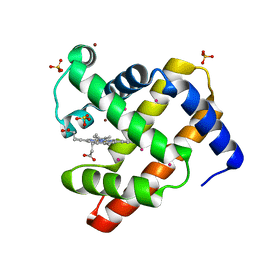 | | SPERM WHALE MYOGLOBIN UNDER XENON PRESSURE 30 Bar | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Colloc'h, N, Prange, T, Vallone, B. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Studies with Xenon and Nitrous Oxide Provide Evidence for Protein-dependent Processes in the Mechanisms of General Anesthesia
Anesthesiology, 121, 2014
|
|
4NEQ
 
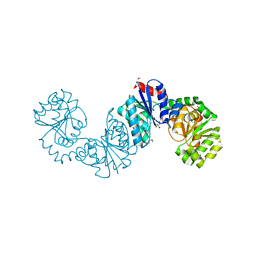 | | The structure of UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of the archaeal UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii reveal a conformational change induced by UDP-GlcNAc.
Proteins, 82, 2014
|
|
3FND
 
 | |
3H2S
 
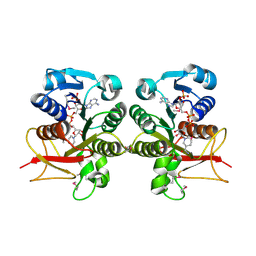 | | Crystal Structure of the Q03B84 Protein from Lactobacillus casei. Northeast Structural Genomics Consortium Target LcR19. | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NADH-flavin reductase | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J, Nair, R, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the Q03B84 protein from Lactobacillus casei.
To be Published
|
|
1P5E
 
 | | The structure of phospho-CDK2/cyclin A in complex with the inhibitor 4,5,6,7-tetrabromobenzotriazole (TBS) | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, Cell division protein kinase 2, Cyclin A2 | | Authors: | De Moliner, E, Brown, N.R, Johnson, L.N. | | Deposit date: | 2003-04-26 | | Release date: | 2003-07-01 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Alternative binding modes of an inhibitor to two different kinases
Eur.J.Biochem., 270, 2003
|
|
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
1S16
 
 | | Crystal Structure of E. coli Topoisomerase IV ParE 43kDa subunit complexed with ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
4PSH
 
 | | Structure of holo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3, ARGININE | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
