8BT0
 
 | | Notum Inhibitor ARUK3005518 | | Descriptor: | (3~{a}~{S})-2,2-bis(fluoranyl)-3~{a},4-dihydro-3~{H}-pyrrolo[1,2-a]indol-1-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTC
 
 | | Notum Inhibitor ARUK3004558 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4,5-bis(chloranyl)-2,3-dihydroindol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTI
 
 | | Notum Inhibitor ARUK3004556 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chloranylindol-1-yl)-2-methoxy-ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSZ
 
 | | Notum Inhibitor ARUK3005522 | | Descriptor: | (3~{a}~{R})-2,3,3~{a},4-tetrahydropyrrolo[1,2-a]indol-1-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTA
 
 | | Notum Inhibitor ARUK3004308 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chloranyl-2,3-dihydroindol-1-yl)ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTH
 
 | | Notum Inhibitor ARUK3004552 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydroindol-1-yl)prop-2-en-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT8
 
 | | Notum Inhibitor ARUK3004048 | | Descriptor: | 1,2-ETHANEDIOL, 1-indol-1-ylethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
7A1R
 
 | |
6PEG
 
 | | MIF with a allosteric inhibitor | | Descriptor: | 4-amino-5-hydroxy-6-[(E)-(3-{[3-(2-methylpropanoyl)pyrazolo[1,5-a]pyridin-2-yl]methyl}phenyl)diazenyl]naphthalene-1,3-disulfonic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Macrophage Migration Inhibitory Factor by a Chimera of Two Allosteric Binders.
Acs Med.Chem.Lett., 11, 2020
|
|
7QO8
 
 | |
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
6PYD
 
 | | Structure of 3E9 antibody Fab bound to marinobufagenin | | Descriptor: | (3beta,5beta,14alpha,15beta)-3,5-dihydroxy-14,15-epoxybufa-20,22-dienolide, 3E9 anti-marinobufagenin antibody Fab heavy chain, recloned with human IgG4 C region, ... | | Authors: | Franklin, M.C, Macdonald, L.E, McWhirter, J, Murphy, A.J. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kappa-on-Heavy (KoH) bodies are a distinct class of fully-human antibody-like therapeutic agents with antigen-binding properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8UWM
 
 | | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, borolane-based compound Q41 bound | | Descriptor: | 2-methyl-4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzene-1-sulfonamide, CALCIUM ION, Fluorophosphonate-binding serine hydrolase E | | Authors: | Fellner, M. | | Deposit date: | 2023-11-06 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of covalent inhibitors of Staphylococcus aureus serine hydrolases important for virulence and biofilm formation.
Nat Commun, 16, 2025
|
|
6G7R
 
 | | Structure of fully reduced variant E28Q of E. coli hydrogenase-1 at pH 8 | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GAM
 
 | | Structure of E14Q variant of E. coli hydrogenase-2 (as-isolated enzyme) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GUS
 
 | |
6ZG8
 
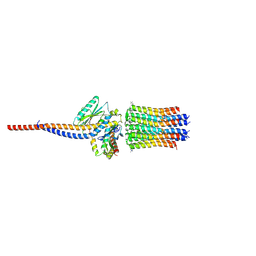 | | bovine ATP synthase rotor domain state 2 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-18 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7QYN
 
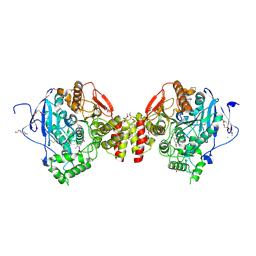 | | Mus musculus acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R2F
 
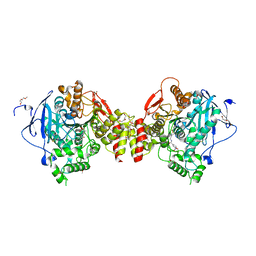 | | Structure of tabun inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
6GW6
 
 | |
7PUL
 
 | | Crystal structure of Endoglycosidase E GH20 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, GLY-ALA-GLY-ALA-ALA, ... | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
6OJT
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 LsdA phenylazophenol complex | | Descriptor: | 4-Hydroxyazobenzene, FE (III) ION, Lignostilbene-alpha,beta-dioxygenase isozyme I | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
7B5G
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2020-12-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
6HCK
 
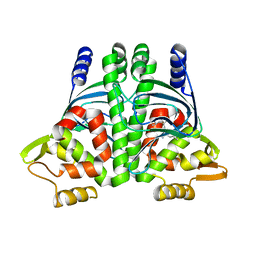 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with dipeptide Leu-Leu | | Descriptor: | LEUCINE, Listeriolysin regulatory protein, SODIUM ION | | Authors: | Grundstrom, C, Oelker, M, Krypotou, E, Scortti, M, Luisi, B.F, Vazquez-Boland, J, Sauer-Eriksson, A.E. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of Bacterial Virulence through the Peptide Signature of the Habitat.
Cell Rep, 26, 2019
|
|
7NPR
 
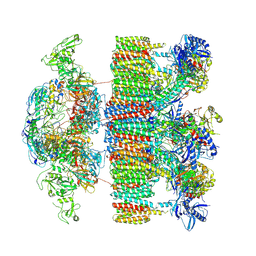 | | Structure of an intact ESX-5 inner membrane complex, Composite C3 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
