6RJ5
 
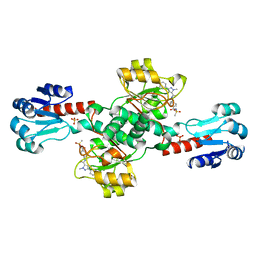 | | Crystal structure of PHGDH in complex with compound 39 | | Descriptor: | 2-methyl-~{N}-[(1~{R})-1-[4-(methylsulfonylcarbamoyl)phenyl]ethyl]-5-phenyl-pyrazole-3-carboxamide, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5LZG
 
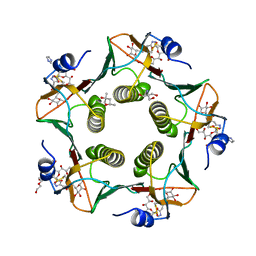 | | Cholera toxin El Tor B-pentamer in complex with inhibitor PC262 | | Descriptor: | (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Heggelund, J.E, Martinsen, T, Krengel, U. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Towards new cholera prophylactics and treatment: Crystal structures of bacterial enterotoxins in complex with GM1 mimics.
Sci Rep, 7, 2017
|
|
6ZT5
 
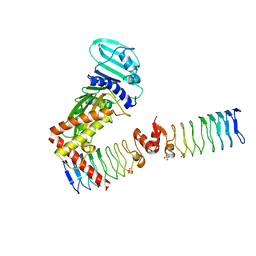 | | Complex between a homodimer of Mycobacterium smegmatis MfpA and a single copy of the N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit | | Descriptor: | DNA gyrase subunit B, Pentapeptide repeat protein MfpA, SULFATE ION | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6RJZ
 
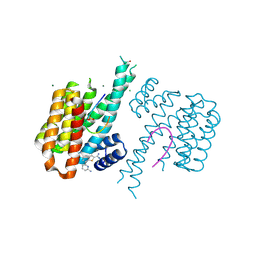 | | Fragment AZ-015 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(6-azanylpyridin-2-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RK5
 
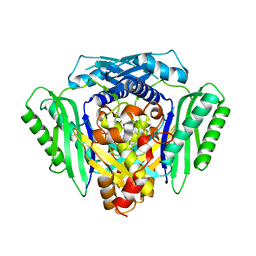 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RL0
 
 | | Recombinant Pseudomonas stutzeri nitrous oxide reductase, form I | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Wuest, A, Prasser, B, Mueller, C, Einsle, O. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Functional assembly of nitrous oxide reductase provides insights into copper site maturation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5M5B
 
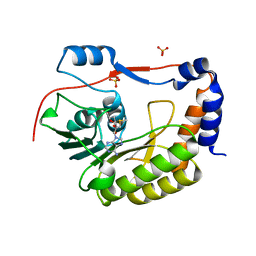 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
7A6O
 
 | |
6ZT3
 
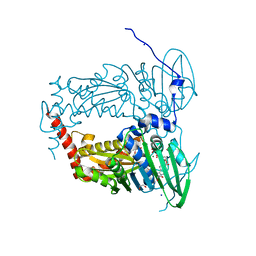 | | N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit complexed with ADPNP | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZT4
 
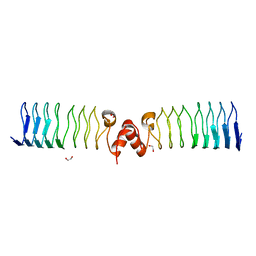 | | Pentapeptide repeat protein MfpA from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Pentapeptide repeat protein MfpA | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6RSJ
 
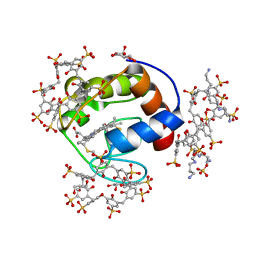 | |
6RO8
 
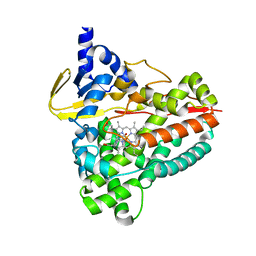 | | The crystal structure of Acinetobacter radioresistens CYP116B5 heme domain | | Descriptor: | Cytochrome P450 RhF, HISTIDINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ciaramella, A, Catucci, G, Gilardi, G, Di Nardo, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of bacterial CYP116B5 heme domain: New insights on class VII P450s structural flexibility and peroxygenase activity.
Int.J.Biol.Macromol., 140, 2019
|
|
6RSP
 
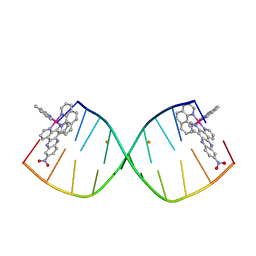 | |
7A6P
 
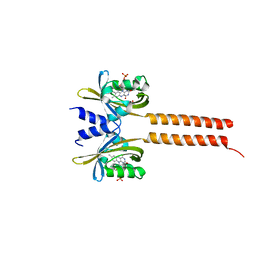 | |
6RKM
 
 | | Fragment AZ-022 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-phenyl-5-phenylazanyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5M4F
 
 | | Complex structure of human protein kinase CK2 catalytic subunit with the inhibitor 4'-carboxy-6,8-chloro-flavonol (FLC21) crystallized under low-salt conditions | | Descriptor: | 4-[6,8-bis(chloranyl)-3-oxidanyl-4-oxidanylidene-chromen-2-yl]benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.519 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
7A6X
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 56 | | Descriptor: | (2S,9S,12R)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-24,27-dimethoxy-11,18,22-trioxa-4-azatetracyclo[21.2.2.113,17.04,9]octacosa-1(25),13(28),14,16,23,26-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AD6
 
 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
7A0D
 
 | | The Crystal Structure of Bovine Thrombin in complex with Hirudin (C16U/C28U) at 1.6 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-1, Prothrombin, ... | | Authors: | Hidmi, T, Mousa, R, Pomyalov, S, Lansky, S, Khouri, L, Metanis, N, Shoham, G. | | Deposit date: | 2020-08-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diselenide crosslinks for enhanced and simplified oxidative protein folding
Commun Chem, 4, 2021
|
|
7AD7
 
 | | Crystal structure of human complement C5 in complex with the K8 bovine knob domain peptide. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
5M1N
 
 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Manganese | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, Phage terminase large subunit, ... | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
6RL4
 
 | | Fragment AZ-025 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[1-(2-azanylethyl)imidazol-4-yl]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RR4
 
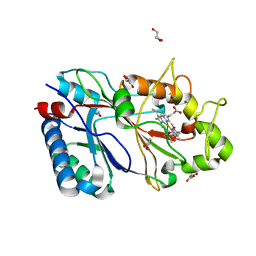 | | Structure of 25% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
6RRK
 
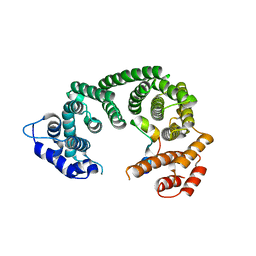 | | Crystal structure of the central region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
