4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V5I
 
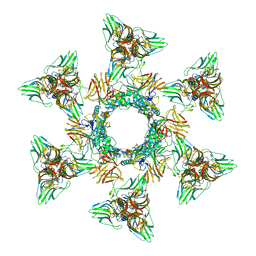 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4UD1
 
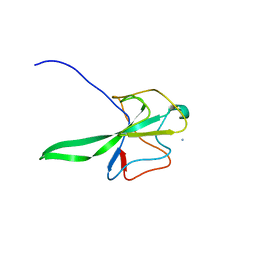 | | Structure of the N Terminal domain of the MERS CoV nucleocapsid | | Descriptor: | AMMONIUM ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Papageorgiou, N, Lichiere, J, Ferron, F, Canard, B, Coutard, B. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Characterization of the N-Terminal Part of the Mers-Cov Nucleocapsid by X-Ray Diffraction and Small-Angle X-Ray Scattering
Acta Crystallogr.,Sect.D, 72, 2016
|
|
8RCQ
 
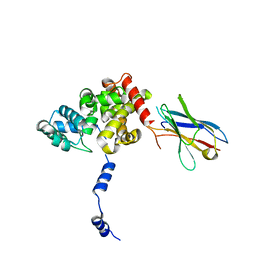 | | Structural flexibility of Nucleoprotein of the Toscana virus in the presence of a nanobody. | | Descriptor: | Nucleoprotein, VHH | | Authors: | Papageorgiou, N, Ferron, F, Coutard, B, Lichiere, J, Baklouti, A. | | Deposit date: | 2023-12-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural flexibility of Toscana virus nucleoprotein in the presence of a single-chain camelid antibody.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5FVA
 
 | | Toscana Virus Nucleocapsid Protein | | Descriptor: | NUCLEOPROTEIN, SODIUM ION | | Authors: | Baklouti, A, Sevajol, M, Goulet, A, Lichiere, J, Ferron, F, Canard, B, Charrel, R.N, Coutard, B, Papageorgiou, N. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Toscana virus nucleoprotein oligomer organization observed in solution.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5M5B
 
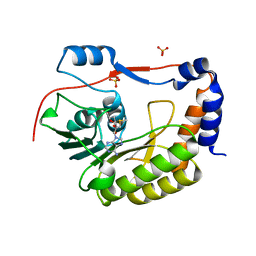 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
6G13
 
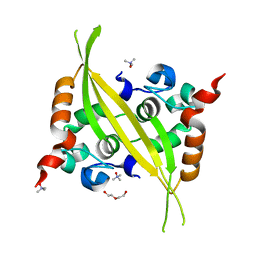 | | C-terminal domain of MERS-CoV nucleocapsid | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Nguyen, T.H.V, Ferron, F.P, Lichiere, J, Canard, B, Papageorgiou, N, Coutard, B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3U6X
 
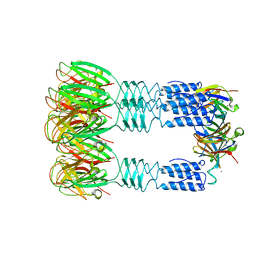 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UH8
 
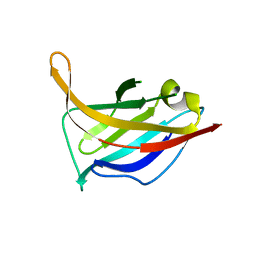 | | N-terminal domain of phage TP901-1 ORF48 | | Descriptor: | ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3HG0
 
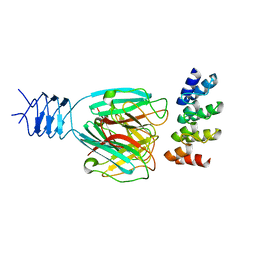 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
3ILD
 
 | | Structure of ORF157-K57A from Acidianus filamentous virus 1 | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
3ILE
 
 | | Crystal structure of ORF157-E86A of Acidianus filamentous virus 1 | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
2X53
 
 | | Structure of the phage p2 baseplate in its activated conformation with Sr | | Descriptor: | ORF15, ORF16, PUTATIVE RECEPTOR BINDING PROTEIN, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2X8K
 
 | | Crystal Structure of SPP1 Dit (gp 19.1) Protein, a Paradigm of Hub Adsorption Apparatus in Gram-positive Infecting Phages. | | Descriptor: | HYPOTHETICAL PROTEIN 19.1 | | Authors: | Veesler, D, Robin, G, Lichiere, J, Auzat, I, Tavares, P, Bron, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Bacteriophage Spp1 Distal Tail Protein (Gp 19.1): A Baseplate Hub Paradigm in Gram+ Infecting Phages.
J.Biol.Chem., 285, 2010
|
|
2WZP
 
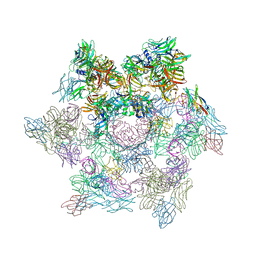 | | Structures of Lactococcal Phage p2 Baseplate Shed Light on a Novel Mechanism of Host Attachment and Activation in Siphoviridae | | Descriptor: | CAMELID VHH5, LACTOCOCCAL PHAGE P2 ORF15, LACTOCOCCAL PHAGE P2 ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XF6
 
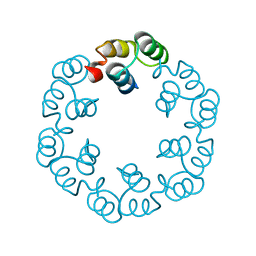 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XF5
 
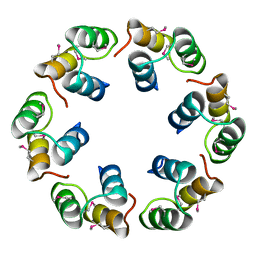 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XF7
 
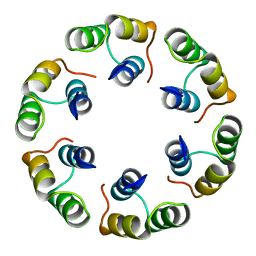 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. High-resolution structure. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
3D8L
 
 | | Crystal structure of ORF12 from the lactococcus lactis bacteriophage p2 | | Descriptor: | ORF12 | | Authors: | Siponen, M.I, Spinelli, S, Lichiere, J, Moineau, S, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-05-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ORF12 from Lactococcus lactis phage p2 identifies a tape measure protein chaperone
J.Bacteriol., 191, 2009
|
|
3EJC
 
 | | Full length Receptor Binding Protein from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein (BPP) | | Authors: | Spinelli, S, Lichiere, J, Blangy, S, Sciara, G, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
J.Biol.Chem., 285, 2010
|
|
