1GH7
 
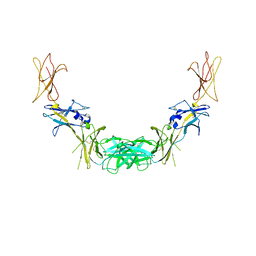 | | CRYSTAL STRUCTURE OF THE COMPLETE EXTRACELLULAR DOMAIN OF THE BETA-COMMON RECEPTOR OF IL-3, IL-5, AND GM-CSF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOKINE RECEPTOR COMMON BETA CHAIN, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carr, P.D, Gustin, S.E, Church, A.P, Murphy, J.M, Ford, S.C, Mann, D.A, Woltring, D.M, Walker, I, Ollis, D.L, Young, I.G. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration.
Cell(Cambridge,Mass.), 104, 2001
|
|
5IRE
 
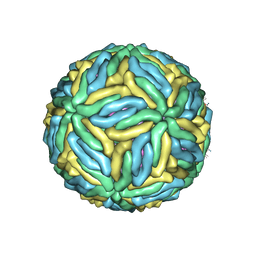 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
7FJC
 
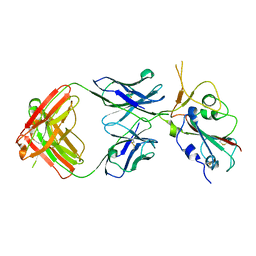 | | Crystal structure of SARS-CoV-2 Beta RBD complexed with P36-5D2 Fab | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike protein S1, ... | | Authors: | Zhang, L.Q, Wang, X.Q, Shan, S.S, Lan, J. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of SARS-CoV-2 Beta RBD complexed with P36-5D2 Fab
To Be Published
|
|
7KE4
 
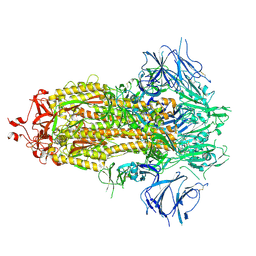 | |
7UGN
 
 | |
7UWN
 
 | | Structure of the IL-17A-IL-17RA-IL-17RC ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7KE6
 
 | |
7UGO
 
 | |
5FC5
 
 | | Murine SMPDL3A in complex with phosphocholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
5A6Y
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with mannose-alpha1,3mannoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, GLYCEROL, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
6JEQ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with beta-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7UAB
 
 | | Human pro-meprin alpha (zymogen state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7KYA
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7UAI
 
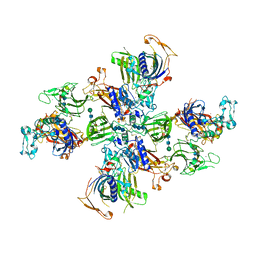 | | Meprin alpha helix in complex with fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
6HTL
 
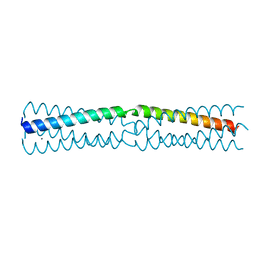 | |
7UAC
 
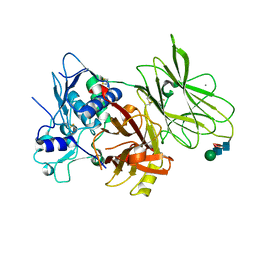 | | Human pro-meprin alpha (zymogen state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Meprin A subunit alpha, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7UOV
 
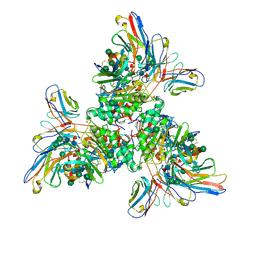 | |
7UAF
 
 | | Human meprin alpha inhibitory complex with compound 10d (N~3~,N~3~-bis[(2H-1,3-benzodioxol-5-yl)methyl]-N-hydroxy-beta-alaninamide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
7KEC
 
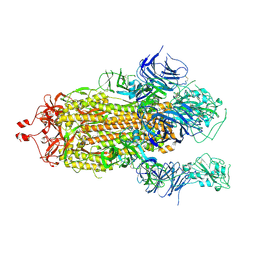 | |
7KDJ
 
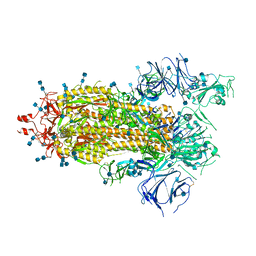 | |
7UAE
 
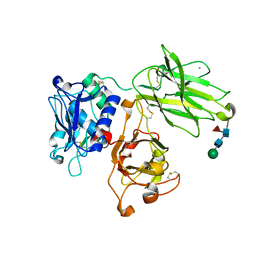 | | Human meprin alpha (active state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bayly-Jones, C, Lupton, C.J, Fritz, C, Schlenzig, D, Whisstock, J.C. | | Deposit date: | 2022-03-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Helical ultrastructure of the metalloprotease meprin alpha in complex with a small molecule inhibitor.
Nat Commun, 13, 2022
|
|
1GAC
 
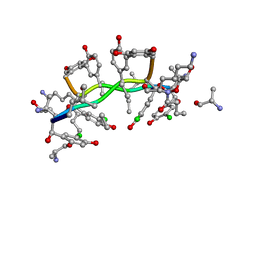 | | NMR structure of asymmetric homodimer of a82846b, a glycopeptide antibiotic, complexed with its cell wall pentapeptide fragment | | Descriptor: | CELL WALL PENTAPEPTIDE, CHLOROORIENTICIN A, vancosamine, ... | | Authors: | Kline, A.D, Prowse, W.G, Skelton, M.A, Loncharich, R.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformation of A82846B, a Glycopeptide Antibiotic, Complexed with its Cell Wall Fragment: An Asymmetric Homodimer Determined Using NMR Spectroscopy.
Biochemistry, 34, 1995
|
|
7UOT
 
 | | Native Lassa glycoprotein in complex with neutralizing antibodies 8.9F and 37.2D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 37.2D heavy chain (variable domain), ... | | Authors: | Li, H, Saphire, E.O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | A cocktail of protective antibodies subverts the dense glycan shield of Lassa virus.
Sci Transl Med, 14, 2022
|
|
4ZZT
 
 | | Geotrichum candidum Cel7A structure complex with thio-linked cellotriose at 1.56A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL7A, GLYCEROL, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
4ZZW
 
 | | Geotrichum candidum Cel7A structure complex with cellobiose at 1.5A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE I, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
