5CVS
 
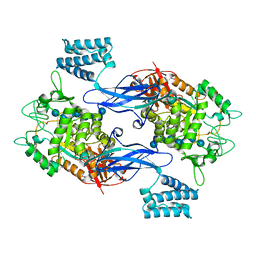 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltoheptaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Rashid, A.M, Syson, K, Koliwer-Brandl, H, van de Weerd, R, Stevenson, C.E.M, Batey, S.F.D, Miah, F, Alber, M, Ioerger, T.R, Chandra, G, Appelmelk, B.J, Nartowski, K.P, Khimyak, Y.Z, Lawson, D.M, Jacobs, W.R, Geurtsen, J, Kalscheuer, R, Bornemann, S. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
6XV9
 
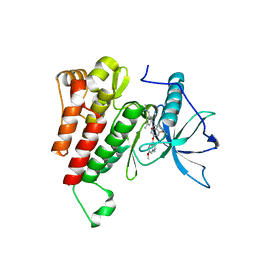 | | Crystal structure of the kinase domain of human c-KIT in complex with a type-II inhibitor | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-[3-[(dimethylamino)methyl]-5-methyl-phenyl]-2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanamide | | Authors: | Ogg, D.J, Howard, T, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
8Z4L
 
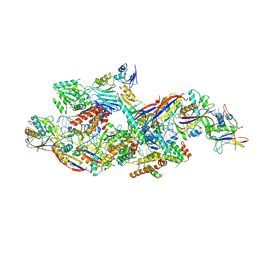 | |
8Z9E
 
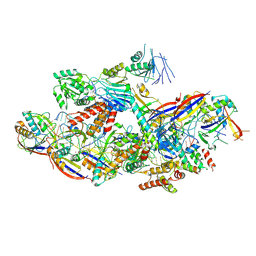 | |
8Z9C
 
 | |
8Z99
 
 | |
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6R5H
 
 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
9BHN
 
 | |
9C6Q
 
 | |
3GZ9
 
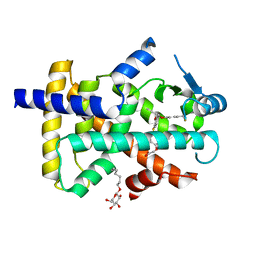 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd) in Complex with a Full Agonist | | Descriptor: | (2,3-dimethyl-4-{[2-(prop-2-yn-1-yloxy)-4-{[4-(trifluoromethyl)phenoxy]methyl}phenyl]sulfanyl}phenoxy)acetic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6XSZ
 
 | |
4V8J
 
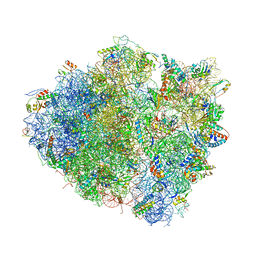 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2011-12-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7EXA
 
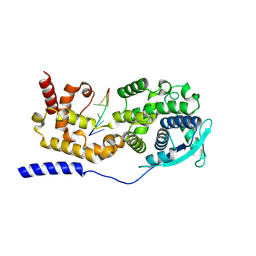 | | Structure of mumps virus nucleoprotein without C-arm | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N, Qin, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
6XVA
 
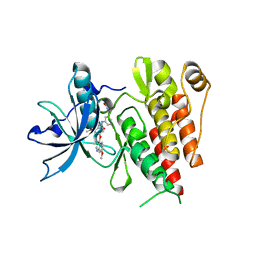 | | Crystal structure of the kinase domain of human c-KIT in complex with a type-II inhibitor bearing an acrylamide | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-[[3-[2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanoylamino]-5-methyl-phenyl]methyl]propanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
8VJW
 
 | |
8JJP
 
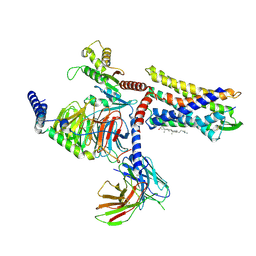 | | G protein-coupled receptor 1 | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, A, Liu, Y, Chen, G, Ye, F. | | Deposit date: | 2023-05-31 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | G protein-coupled receptor 1
To Be Published
|
|
8RZX
 
 | |
9J1T
 
 | | Structure of a triple-helix region of human Collagen type IV from Trautec | | Descriptor: | GLYCEROL, Triple-helix region of human collagen type IV | | Authors: | Fan, X, Chu, Y, Zhai, Y, Fu, S, Li, D, Feng, P, Cao, K, Wu, X, Cai, H, Wang, H, Qian, S. | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type IV from Trautec
To Be Published
|
|
9J23
 
 | | Structure of a triple-helix region of human Collagen type II from Trautec | | Descriptor: | Triple-helix region of human collagen type II | | Authors: | Fan, X, Zhai, Y, Chu, Y, Fu, S, Li, D, Feng, P, Cao, K, Li, J, Si, Y, Ma, L, Qian, S. | | Deposit date: | 2024-08-06 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
8UR8
 
 | |
5HJO
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with bound substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-alpha-D-arabino-hexopyranose-(1-3)-D-glucal, ACETATE ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-01-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8YDC
 
 | | Crystal structure of a hammerhead ribozyme with pseudoknot | | Descriptor: | DNA/RNA (5'-R(*AP*CP*AP*UP*GP*UP*CP*U)-D(P*C)-R(P*UP*GP*GP*GP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, ribozyme strand | | Authors: | Liu, Y, Zhan, X. | | Deposit date: | 2024-02-20 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure and catalytic mechanism of a pseudoknot-containing hammerhead ribozyme.
Nat Commun, 15, 2024
|
|
8Z4J
 
 | |
9B6I
 
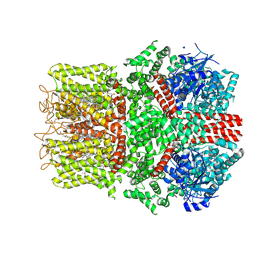 | | Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014 | | Descriptor: | 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
