5T1S
 
 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5C0A
 
 | | 1E6 TCR in complex with HLA-A02 carrying MVW peptide | | Descriptor: | 1,2-ETHANEDIOL, 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5T29
 
 | | Crystal structure of 10E8 Fab light chain mutant3, against the MPER region of the HIV-1 Env, in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, Antibody 10E8 FAB LIGHT CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
6EMT
 
 | |
9D3W
 
 | | CRYSTAL STRUCTURE OF EGFR(L858R/T790M/C797S) IN COMPLEX WITH AUR-8250 | | Descriptor: | (4S)-1-(1,4-dioxaspiro[4.5]decan-8-yl)-N-{2-[(3S,4R)-3-fluoro-4-methoxypiperidin-1-yl]pyrimidin-4-yl}-2-methyl-1H-imidazo[1,2-b]pyrazol-6-amine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Bell, J.A. | | Deposit date: | 2024-08-12 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of a Novel Mutant-Selective Epidermal Growth Factor Receptor Inhibitor Using an In Silico Enabled Drug Discovery Platform.
J.Med.Chem., 67, 2024
|
|
8BE6
 
 | | Crystal structure of SOS1-HRas-peptidomimetic2 | | Descriptor: | GTPase HRas, SOS1-HRas-peptidomimetic2, Son of sevenless homolog 1 | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89880252 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2VGP
 
 | | Crystal structure of Aurora B kinase in complex with a aminothiazole inhibitor | | Descriptor: | 4-[(5-bromo-1,3-thiazol-2-yl)amino]-N-methylbenzamide, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Andersen, C.B, Wan, Y, Chang, J.W, Lee, C, Liu, Y, Sessa, F, Villa, F, Nallan, L, Musacchio, A, Gray, N.S. | | Deposit date: | 2007-11-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Selective Aminothiazole Aurora Kinase Inhibitors
Acs Chem.Biol., 3, 2008
|
|
5L64
 
 | |
6ROZ
 
 | |
8BE8
 
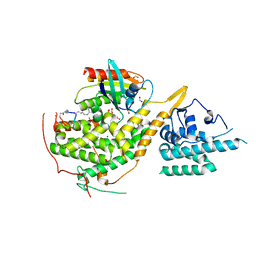 | | Crystal structure of SOS1-HRas-peptidomimetic4 | | Descriptor: | FORMIC ACID, GTPase HRas, SOS1-HRas-peptidomimetic4, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BE7
 
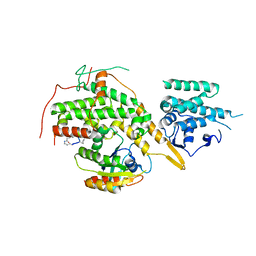 | |
5A2R
 
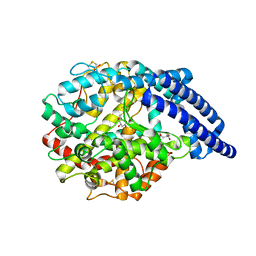 | | A New Crystal Structure of the Drosophila melanogaster Angiotensin Converting Enzyme Homologue AnCE. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Harrison, C, Acharya, K.R. | | Deposit date: | 2015-05-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A New High-Resolution Crystal Structure of the Drosophila Melanogaster Angiotensin Converting Enzyme Homologue, Ance.
FEBS Open Bio, 5, 2015
|
|
8BE9
 
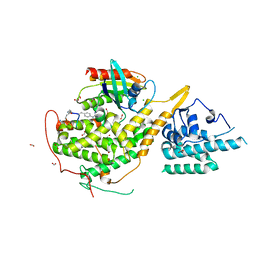 | | Crystal structure of SOS1-HRas-peptidomimetic5 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GTPase HRas, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BEA
 
 | |
1KIY
 
 | | D100E trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, trichodiene synthase | | Authors: | Rynkiewicz, M.J, Cane, D.E, Christianson, D.W. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structures of D100E trichodiene synthase and its pyrophosphate complex reveal the basis for terpene product diversity.
Biochemistry, 41, 2002
|
|
8HCJ
 
 | | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans | | Descriptor: | CALCIUM ION, Xylan 1,4-beta-xylosidase | | Authors: | Vishwakarma, P, Sachdeva, E, Goyal, A, Ethayathulla, A.S, Das, U, Kaur, P. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | Deciphering the structural and biochemical aspects of xylosidase from Pseudopedobacter saltans.
Int.J.Biol.Macromol., 291, 2024
|
|
5Z7A
 
 | | Crystal structure of NDP52 SKICH region | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q. | | Deposit date: | 2018-01-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
5E2O
 
 | |
5MC7
 
 | |
9HC2
 
 | | Crystal structure of Lysozyme in complex with Gentisic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dihydroxybenzoic acid, Lysozyme C | | Authors: | Ifeagwu, M.C, Flood, R.J, Mockler, N.M, Crowley, P.B. | | Deposit date: | 2024-11-08 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Lysozyme in complex with Gentisic Acid
To Be Published
|
|
6C5N
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
5A6K
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-NGT | | Descriptor: | (2S,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, GH20C | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Second Beta-Hexosaminidase Encoded in the Streptococcus Pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
J.Biol.Chem., 290, 2015
|
|
7USH
 
 | | BRD2-BD2 in complex with SF2523 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 2 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
8XVM
 
 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
