6C0T
 
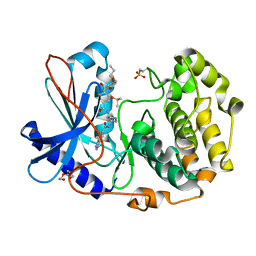 | | Crystal structure of cGMP-dependent protein kinase Ialpha (PKG Ialpha) catalytic domain bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, TRIETHYLENE GLYCOL, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
5MPS
 
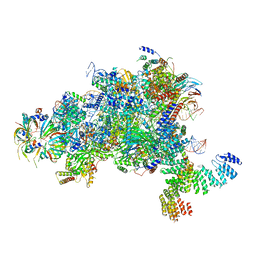 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
6EWI
 
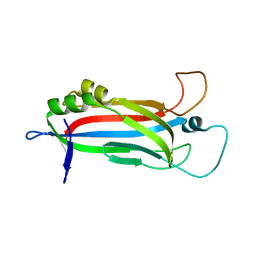 | |
6EWP
 
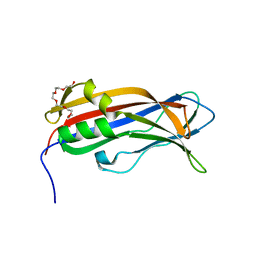 | | Mus musculus CEP120 third C2 domain (C2C) | | Descriptor: | Centrosomal protein of 120 kDa, HEXAETHYLENE GLYCOL | | Authors: | van Breugel, M, al-Jassar, C. | | Deposit date: | 2017-11-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Disease-Associated Mutations in CEP120 Destabilize the Protein and Impair Ciliogenesis.
Cell Rep, 23, 2018
|
|
5K5W
 
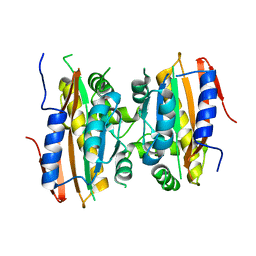 | | Crystal structure of limiting CO2-inducible protein LCIB | | Descriptor: | ZINC ION, limiting CO2-inducible protein LCIB | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1ZG3
 
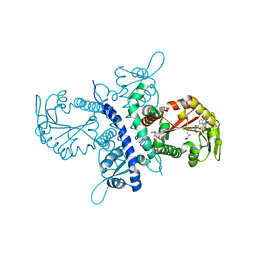 | | Crystal structure of the isoflavanone 4'-O-methyltransferase complexed with SAH and 2,7,4'-trihydroxyisoflavanone | | Descriptor: | (2S,3R)-2,7-DIHYDROXY-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-4H-CHROMEN-4-ONE, S-ADENOSYL-L-HOMOCYSTEINE, isoflavanone 4'-O-methyltransferase | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
6EXN
 
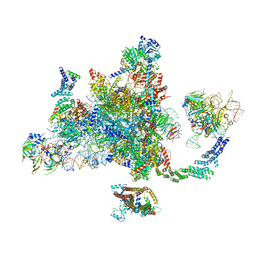 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
6EWH
 
 | |
6EWL
 
 | |
6C0U
 
 | | Crystal structure of cAMP-dependent protein kinase Calpha subunit bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, PHOSPHATE ION, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
1ZHF
 
 | | Crystal structure of selenomethionine substituted isoflavanone 4'-O-methyltransferase | | Descriptor: | Isoflavanone 4'-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
5UCI
 
 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | (2,4-Dihydroxy-3-(hydroxymethyl)-5-isopropylphenyl)(isoindolin-2-yl)methanone, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Peng, S, Balch, M, Matts, R, Deng, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
1MCT
 
 | | THE REFINED 1.6 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PORCINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, TRYPSIN INHIBITOR A | | Authors: | Huang, Q, Liu, S, Tang, Y. | | Deposit date: | 1992-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined 1.6 A resolution crystal structure of the complex formed between porcine beta-trypsin and MCTI-A, a trypsin inhibitor of the squash family. Detailed comparison with bovine beta-trypsin and its complex.
J.Mol.Biol., 229, 1993
|
|
2J3Q
 
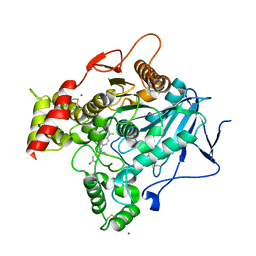 | | Torpedo acetylcholinesterase complexed with fluorophore thioflavin T | | Descriptor: | 2-[4-(DIMETHYLAMINO)PHENYL]-6-HYDROXY-3-METHYL-1,3-BENZOTHIAZOL-3-IUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Harel, M, Cusack, B, Johnson, J.L, Silman, I, Sussman, J.L, Rosenberry, T.L. | | Deposit date: | 2006-08-23 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site.
J. Am. Chem. Soc., 130, 2008
|
|
6F1U
 
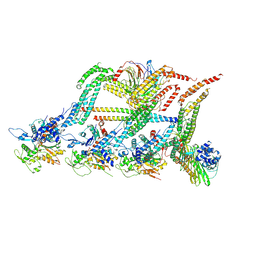 | | N terminal region of dynein tail domains in complex with dynactin filament and BICDR-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARP1 actin related protein 1 homolog A, BICD family-like cargo adapter 1, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F38
 
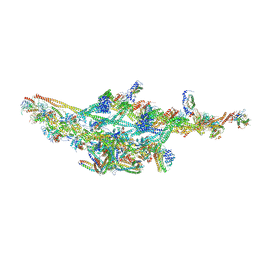 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
5MHQ
 
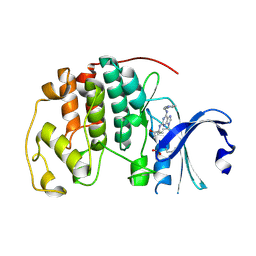 | | CCT068127 in complex with CDK2 | | Descriptor: | (2~{R},3~{S})-3-[[9-propan-2-yl-6-(pyridin-3-ylmethylamino)purin-2-yl]amino]pentan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Whittaker, S.R, Barlow, C, Martin, M.P, Mancusi, C, Wagner, S, Barrie, E, te Poele, R, Sharp, S, Brown, N, Wilson, S, Clarke, P, Walton, M.I, MacDonald, E, Blagg, J, Noble, M.E.M, Garrett, M.D, Workman, P. | | Deposit date: | 2016-11-25 | | Release date: | 2017-12-20 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor.
Mol Oncol, 12, 2018
|
|
4IUY
 
 | | Crystal structure of short-chain dehydrogenase/reductase (apo-form) from A. baumannii clinical strain WM99C | | Descriptor: | Short chain dehydrogenase/reductase | | Authors: | Shah, B.S, Tetu, S.G, Harrop, S.J, Paulsen, I.T, Mabbutt, B.C. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure of a short-chain dehydrogenase/reductase (SDR) within a genomic island from a clinical strain of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
2JF0
 
 | | Mus musculus acetylcholinesterase in complex with tabun and Ortho-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
6F28
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2'-Amino-2'-carboxyethyl]-6-methyl-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.4A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
5N1Q
 
 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTHERMOCOCCUS THERMOLITHOTROPHICUS AT 1.9 A RESOLUTION | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
2JGE
 
 | |
2JEY
 
 | | Mus musculus acetylcholinesterase in complex with HLo-7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
2JEZ
 
 | | Mus musculus acetylcholinesterase in complex with tabun and HLo-7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
