1RYE
 
 | | Crystal Structure of the Shifted Form of the Glucose-Fructose Oxidoreductase from Zymomonas mobilis | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Arora, M, Straza, M, Donnelly, M, Joachimiak, A. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Shifted Form of the Glucose-Fructose Oxidoreductase from Zymomonas mobilis
To be Published
|
|
2YPL
 
 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | AGA T-CELL RECEPTOR ALPHA CHAIN, AGA T-CELL RECEPTOR BETA CHAIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, van der Merwe, P.A, Easterbrook, P, McMichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1RYD
 
 | | Crystal Structure of Glucose-Fructose Oxidoreductase from Zymomonas mobilis | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Arora, M, Straza, M, Joachimiak, A. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glucose-Fructose Oxidoreductase from Zymomonas mobilis
To be Published
|
|
1QZQ
 
 | | human Tyrosyl DNA phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase 1 | | Authors: | Raymond, A.C, Rideout, M.C, Staker, B, Hjerrild, K, Burgin Jr, A.B. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Human Tyrosyl-DNA Phosphodiesterase I Catalytic Residues.
J.Mol.Biol., 338, 2004
|
|
1RJ8
 
 | | The crystal structure of TNF family member EDA-A2 | | Descriptor: | ectodysplasin-A isoform EDA-A2 | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Crystal Structure of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity
STRUCTURE, 11, 2003
|
|
1JPY
 
 | | Crystal structure of IL-17F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hymowitz, S.G, Filvaroff, E.H, Yin, J, Lee, J, Cai, L, Risser, P, Maruoka, M, Mao, W, Foster, J, Kelley, R, Pan, G, Gurney, A.L, de Vos, A.M, Starovasnik, M.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding.
EMBO J., 20, 2001
|
|
1RJ7
 
 | | Crystal structure of EDA-A1 | | Descriptor: | Ectodysplasin A | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity.
Structure, 11, 2003
|
|
1D0G
 
 | | CRYSTAL STRUCTURE OF DEATH RECEPTOR 5 (DR5) BOUND TO APO2L/TRAIL | | Descriptor: | APOPTOSIS-2 LIGAND, CHLORIDE ION, DEATH RECEPTOR-5, ... | | Authors: | Hymowitz, S.G, Christinger, H.W, Fuh, G, O'Connell, M.P, Kelley, R.F, Ashkenazi, A, de Vos, A.M. | | Deposit date: | 1999-09-09 | | Release date: | 1999-10-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5.
Mol.Cell, 4, 1999
|
|
1DG6
 
 | | CRYSTAL STRUCTURE OF APO2L/TRAIL | | Descriptor: | APO2L/TNF-RELATED APOPOTIS INDUCING LIGAND (TRAIL), CHLORIDE ION, ZINC ION | | Authors: | Hymowitz, S.G, O'ConnelL, M.P, Ultsch, M.H, de Vos, A.M, Kelley, R.F. | | Deposit date: | 1999-11-23 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A unique zinc-binding site revealed by a high-resolution X-ray structure of homotrimeric Apo2L/TRAIL.
Biochemistry, 39, 2000
|
|
3DLW
 
 | | Antichymotrypsin | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Feil, S.C. | | Deposit date: | 2008-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and characterization of a misfolded monomeric serpin formed at physiological temperature
J.Mol.Biol., 403, 2010
|
|
2LFT
 
 | | Human prion protein with E219K protective polymorphism | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giacin, G, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-07-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism.
Biochem.J., 446, 2012
|
|
5CXO
 
 | | Intriguing role of epoxide hydrolase/cyclase-like enzyme SalBIII in pyran ring formation in polyether salinomycin | | Descriptor: | Epoxide hydrolase, HEXAETHYLENE GLYCOL | | Authors: | Dias, M.V.B, Luhavaya, H, Williams, S.R, Hong, H, Oliveira, L.G, Leadlay, P.F. | | Deposit date: | 2015-07-29 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymology of Pyran Ring A Formation in Salinomycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1JC6
 
 | | SOLUTION STRUCTURE OF BUNGARUS FACIATUS IX, A KUNITZ-TYPE CHYMOTRYPSIN INHIBITOR | | Descriptor: | VENOM BASIC PROTEASE INHIBITORS IX AND VIIIB | | Authors: | Chen, C, Hsu, C.H, Su, N.Y, Chiou, S.H, Wu, S.H. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus
J.BIOL.CHEM., 276, 2001
|
|
4UBY
 
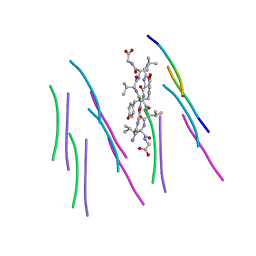 | |
8ZMY
 
 | | F0502B-bound WT polymorph 5a alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6RT0
 
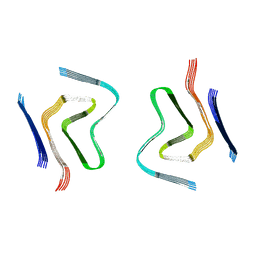 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
8ZLP
 
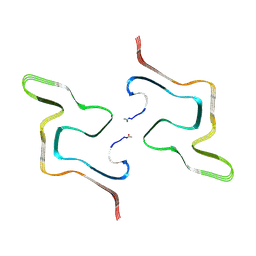 | |
6RTB
 
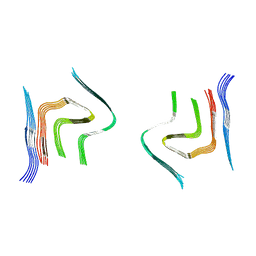 | | cryo-em structure of alpha-synuclein fibril polymorph 2B | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6SSX
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-09-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6SST
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2B | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-09-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
2ZJB
 
 | | Crystal structure of the human Dmc1-M200V polymorphic variant | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Hikiba, J, Hirota, K, Kagawa, W, Ikawa, S, Kinebuchi, T, Sakane, I, Takizawa, Y, Yokoyama, S, Mandon-Pepin, B, Nicolas, A, Shibata, T, Ohta, K, Kurumizaka, H. | | Deposit date: | 2008-03-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional analyses of the DMC1-M200V polymorphism found in the human population
Nucleic Acids Res., 36, 2008
|
|
6CU7
 
 | | Alpha Synuclein fibril formed by full length protein - Rod Polymorph | | Descriptor: | Alpha-synuclein | | Authors: | Li, B, Hatami, A, Ge, P, Murray, K.A, Sheth, P, Zhang, M, Nair, G, Sawaya, M.R, Zhu, C, Broad, M, Shin, W.S, Ye, S, John, V, Eisenberg, D.S, Zhou, Z.H, Jiang, L. | | Deposit date: | 2018-03-23 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel.
Nat Commun, 9, 2018
|
|
6CU8
 
 | | Alpha Synuclein fibril formed by full length protein - Twister Polymorph | | Descriptor: | Alpha-synuclein | | Authors: | Li, B, Hatami, A, Ge, P, Murray, K.A, Sheth, P, Zhang, M, Nair, G, Sawaya, M.R, Zhu, C, Broad, M, Shin, W.S, Ye, S, John, V, Eisenberg, D.S, Zhou, Z.H, Jiang, L. | | Deposit date: | 2018-03-23 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel.
Nat Commun, 9, 2018
|
|
9FYP
 
 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
5JRG
 
 | | Crystal structure of the nucleosome containing the DNA with tetrahydrofuran (THF) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Arimura, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphism of apyrimidinic DNA structures in the nucleosome
Sci Rep, 7, 2017
|
|
