1RA4
 
 | |
8RTN
 
 | | Human thrombin in complex with a trivalent inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, SODIUM ION, ... | | Authors: | Ripoll-Rozada, J, Maxwell, J, Payne, R.J, Pereira, P.J.B. | | Deposit date: | 2024-01-26 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Human thrombin in complex with a trivalent inhibitor
To Be Published
|
|
8T3N
 
 | | Solution NMR structure of synthetic peptide AMPCry10Aa_5 rational designed from Cry10Aa bacterial protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Anti-Staphy Peptides Rationally Designed from Cry10Aa Bacterial Protein.
Acs Omega, 9, 2024
|
|
8T3H
 
 | | Solution NMR structure alpha-helix 3 of Cry10Aa protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix 3 of Cry10Aa protein
To Be Published
|
|
6CBZ
 
 | |
8JZM
 
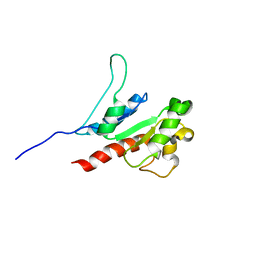 | | The inhibitor of Toll-like receptor signaling o-vanillin binds covalently to MAL/TIRAP Lys-210 | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Rahaman, M.H, Jia, X, Maxwell, M.J, Mobli, M, Kobe, B. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | o-Vanillin binds covalently to MAL/TIRAP Lys-210 but independently inhibits TLR2.
J Enzyme Inhib Med Chem, 39, 2024
|
|
1VJQ
 
 | |
1GUE
 
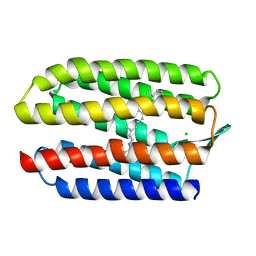 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
1GU8
 
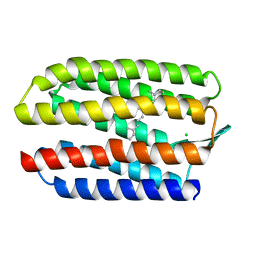 | | SENSORY RHODOPSIN II | | Descriptor: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | Authors: | Edman, K, Royant, A, Nollert, P, Maxwell, C.A, Pebay-Peyroula, E, Navarro, J, Neutze, R, Landau, E.M. | | Deposit date: | 2002-01-24 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor
Structure, 10, 2002
|
|
2STA
 
 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA MAXIMA TRYPSIN INHIBITOR I) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-10 | | Release date: | 2000-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5HJS
 
 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
5HJP
 
 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, DI(HYDROXYETHYL)ETHER, Oxysterols receptor LXR-beta, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
9GU3
 
 | | Human adult muscle nAChR in desensitised state in nanodisc with 1 mM acetylcholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, A, Pike, A.C.W, Chi, G, Webster, R, Maxwell, S, Liu, W, Beeson, D, Sauer, D.B, Dong, Y.Y. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structures of the human adult muscle-type nicotinic receptor in resting and desensitized states.
Cell Rep, 44, 2025
|
|
9GU1
 
 | | Human adult muscle nAChR in resting state in nanodisc with alpha-bungarotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Li, A, Pike, A.C.W, Chi, G, Webster, R, Maxwell, S, Liu, W, Beeson, D, Sauer, D.B, Dong, Y.Y. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structures of the human adult muscle-type nicotinic receptor in resting and desensitized states.
Cell Rep, 44, 2025
|
|
9GU0
 
 | | Human adult muscle nAChR in resting state in detergent with alpha-bungarotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, A, Pike, A.C.W, Chi, G, Webster, R, Maxwell, S, Liu, W, Beeson, D, Sauer, D.B, Dong, Y.Y. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of the human adult muscle-type nicotinic receptor in resting and desensitized states.
Cell Rep, 44, 2025
|
|
9GU2
 
 | | Human adult muscle nAChR in desensitised state in nanodisc with 100 uM acetylcholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, A, Pike, A.C.W, Chi, G, Webster, R, Maxwell, S, Liu, W, Beeson, D, Sauer, D.B, Dong, Y.Y. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structures of the human adult muscle-type nicotinic receptor in resting and desensitized states.
Cell Rep, 44, 2025
|
|
6ARZ
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | BROMIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Calmettes, C, Shah, M, Pawluk, A, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
1LQB
 
 | | Crystal structure of a hydroxylated HIF-1 alpha peptide bound to the pVHL/elongin-C/elongin-B complex | | Descriptor: | Elongin B, Elongin C, Hypoxia-inducible factor 1 ALPHA, ... | | Authors: | Hon, W.C, Wilson, M.I, Harlos, K, Claridge, T.D, Schofield, C.J, Pugh, C.W, Maxwell, P.H, Ratcliffe, P.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2002-05-09 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL.
Nature, 417, 2002
|
|
3V9T
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-N-[(3-ethoxynaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3V9V
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma, methyl 3-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}propanoate | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2011
|
|
1P7R
 
 | | CRYSTAL STRUCTURE OF REDUCED, CO-EXPOSED COMPLEX OF CYTOCHROME P450CAM WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
1P2Y
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM IN COMPLEX WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
2L04
 
 | | The Solution Structure of the C-terminal Ig-like Domain of the Bacteriophage Lambda Tail Tube Protein | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Gasmi-Seabrook, G.M.C, Donaldson, L.W, Howell, P, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the C-Terminal Ig-like Domain of the Bacteriophage l Tail Tube Protein.
J.Mol.Biol., 403, 2010
|
|
2LSM
 
 | | Solution structure of gpFI C-terminal domain | | Descriptor: | DNA-packaging protein FI | | Authors: | Popovic, A, Wu, B, Edwards, A.M, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of phage lambda FI protein (gpFI) reveals a novel mechanism of DNA packaging chaperone activity.
J.Biol.Chem., 287, 2012
|
|
1Z1Z
 
 | |
