2QE5
 
 | |
3BSC
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 5-hydroxy-4-(7-methoxy-1,1-dioxido-2H-1,2,4-benzothiadiazin-3-yl)-2-(3-methylbutyl)-6-phenylpyridazin-3(2H)-one, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5FPS
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 3-aminobenzene-1,2-dicarboxylic acid (AT1246) in an alternate binding site. | | Descriptor: | 3-AMINOBENZENE-1,2-DICARBOXYLIC ACID, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TN2
 
 | | NS5b in complex with lactam-thiophene carboxylic acids | | Descriptor: | 3-[(2R)-2-cyclohexyl-5-oxopyrrolidin-1-yl]-5-phenylthiophene-2-carboxylic acid, Genome polyprotein | | Authors: | Chopra, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of lactam-thiophene carboxylic acids as potent hepatitis C virus polymerase inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5FPT
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site. | | Descriptor: | (1-methyl-1H-indol-3-yl)acetic acid, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Pathuri, P, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4A92
 
 | | Full-length HCV NS3-4A protease-helicase in complex with a macrocyclic protease inhibitor. | | Descriptor: | (1'R,2R,2'S,6S,24AS)-17-FLUORO-6-(1-METHYL-2-OXOPIPERIDINE-3-CARBOXAMIDO)-19,19-DIOXIDO-5,21,24-TRIOXO-2'-VINYL-1,2,3,5,6,7,8,9,10,11,12,13,14,20,21,23,24,24A-OCTADECAHYDROSPIRO[BENZO[S]PYRROLO[2,1-G][1,2,5,8,18]THIATETRAAZACYCLOICOSINE-22,1'-CYCLOPRO-2-CARBOXYLATEPAN]-2-YL 4-FLUOROISOINDOLINE, SERINE PROTEASE NS3, ZINC ION | | Authors: | Schiering, N, D'Arcy, A, Simic, O, Eder, J, Raman, P, Svergun, D.I, Bodendorf, U. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Macrocyclic Hcv Ns3/4A Protease Inhibitor Interacts with Protease and Helicase Residues in the Complex with its Full- Length Target.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4K8B
 
 | | Crystal structure of HCV NS3/4A protease complexed with inhibitor | | Descriptor: | N-(tert-butylcarbamoyl)-3-methyl-L-valyl-(4R)-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-4-[(7-methoxy-2-phenylquinolin-4-yl)oxy]-L-prolinamide, NS3 protease, Nonstructural protein, ... | | Authors: | Nar, H. | | Deposit date: | 2013-04-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand bioactive conformation plays a critical role in the design of drugs that target the hepatitis C virus NS3 protease.
J.Med.Chem., 57, 2014
|
|
4MK7
 
 | |
1NS3
 
 | | STRUCTURE OF HCV PROTEASE (BK STRAIN) | | Descriptor: | NS3 PROTEASE, NS4A PEPTIDE, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
5W2E
 
 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
1CSJ
 
 | | CRYSTAL STRUCTURE OF THE RNA-DEPENDENT RNA POLYMERASE OF HEPATITIS C VIRUS | | Descriptor: | HEPATITIS C VIRUS RNA POLYMERASE (NS5B) | | Authors: | Bressanelli, S, Tomei, L, Roussel, A, Incitti, I, Vitale, R.L, Mathieu, M, De Francesco, R, Rey, F.A. | | Deposit date: | 1999-08-18 | | Release date: | 1999-11-08 | | Last modified: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4MIA
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with RG7109 (N-{4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-2,5-dihydropyridin-3-yl)quinolin-3-yl]phenyl}methanesulfonamide) | | Descriptor: | N-{4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-2,5-dihydropyridin-3-yl)quinolin-3-yl]phenyl}methanesulfonamide, RNA-directed RNA polymerase, ZINC ION | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of N-[4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-1H-pyridin-3-yl)-3-quinolyl]phenyl]methanesulfonamide (RG7109), a potent inhibitor of the hepatitis C virus NS5B polymerase.
J.Med.Chem., 57, 2014
|
|
4B6F
 
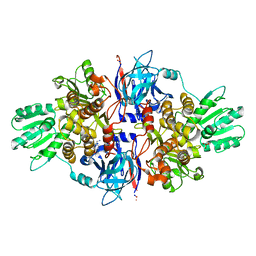 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1NHV
 
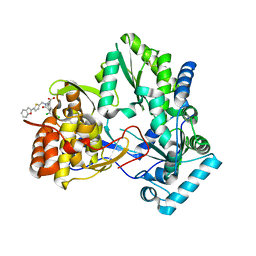 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
2QE2
 
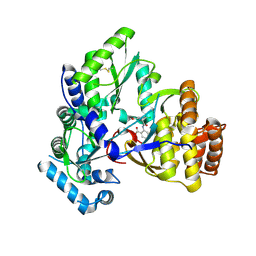 | | Structure of HCV NS5B Bound to an Anthranilic Acid Inhibitor | | Descriptor: | 2-{[N-(2-ACETYL-5-CHLORO-4-FLUOROPHENYL)GLYCYL]AMINO}BENZOIC ACID, RNA-directed RNA polymerase | | Authors: | Chopra, R, Svenson, K, Bard, J. | | Deposit date: | 2007-06-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Anthranilic Acid Derivatives as a Novel Class of Allosteric Inhibitors of Hepatitis C NS5B Polymerase
J.Med.Chem., 50, 2007
|
|
1BT7
 
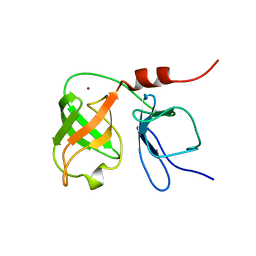 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
