7L31
 
 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, strychnine bound state, 3.8 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-2, Glycine receptor subunit beta,Green fluorescent protein, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
5BKG
 
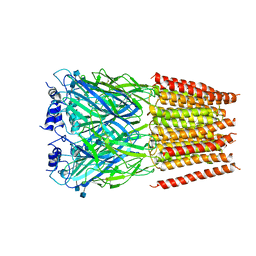 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, glycine bound, (semi)open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alpha-2, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2021-03-19 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
8J1E
 
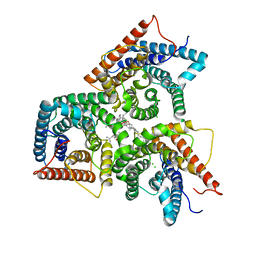 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
6VAL
 
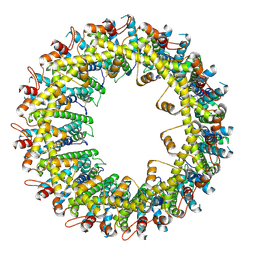 | |
8DN2
 
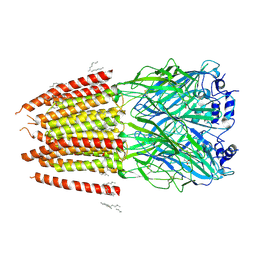 | |
7BTO
 
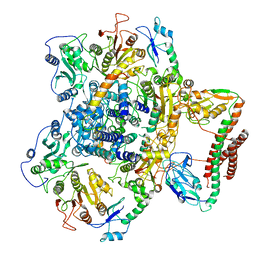 | | EcoR124I-ArdA in the Translocation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTP
 
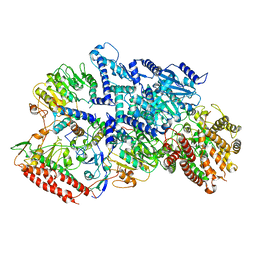 | | EcoR124I-Ocr in Restriction-Alleviation State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7YDQ
 
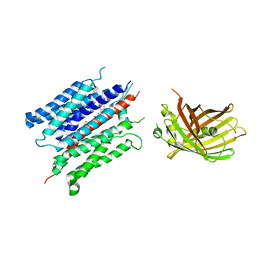 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
8DN4
 
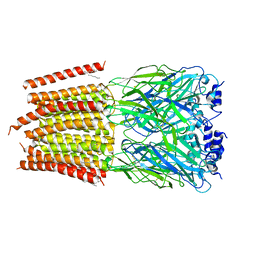 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
8HCO
 
 | | Substrate-engaged TOM complex from yeast | | Descriptor: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | Authors: | Zhou, X.Y, Yang, Y.Q, Wang, G.P, Wang, S.S. | | Deposit date: | 2022-11-02 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7AA5
 
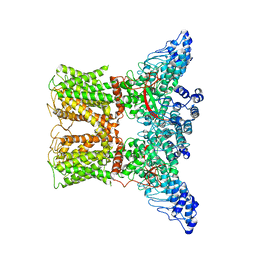 | | Human TRPV4 structure in presence of 4a-PDD | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | Authors: | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
7BST
 
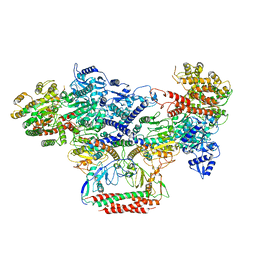 | | EcoR124I-Ocr in the Intermediate State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-03-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTR
 
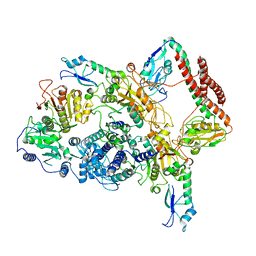 | | EcoR124I-ArdA in the Restriction-Alleviation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTQ
 
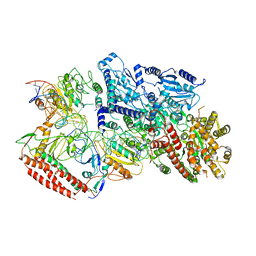 | | EcoR124I-DNA in the Restriction-Alleviation State | | Descriptor: | DNA (64-MER), Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
6MB2
 
 | | Cryo-EM structure of the PYD filament of AIM2 | | Descriptor: | Green fluorescent protein, Interferon-inducible protein AIM2 | | Authors: | Lu, A, Li, Y, Wu, H. | | Deposit date: | 2018-08-29 | | Release date: | 2018-09-05 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2.
Cell Discov, 1, 2015
|
|
7KS0
 
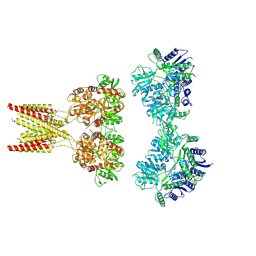 | | GluK2/K5 with 6-Cyano-7-nitroquinoxaline-2,3-dione (CNQX) | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
7KS3
 
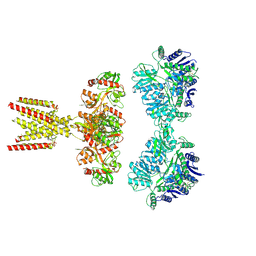 | | GluK2/K5 with L-Glu | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
8FCK
 
 | | Structure of the vertebrate augmin complex | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera, HAUS augmin like complex subunit 4 L homeolog, ... | | Authors: | Travis, S.M, Huang, W, Zhang, R, Petry, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.88 Å) | | Cite: | Integrated model of the vertebrate augmin complex.
Nat Commun, 14, 2023
|
|
