6DV4
 
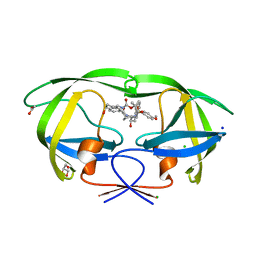 | | HIV-1 wild type protease with GRL-04315A, a tetrahydronaphthalene carboxamide with (R)-Boc-amine and (S)-hydroxyl functionalities as the P2 ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
6DV0
 
 | | HIV-1 wild type protease with GRL-02815A, a thiochroman heterocycle with (S)-Boc-amine functionality as the P2 ligand | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
6DIF
 
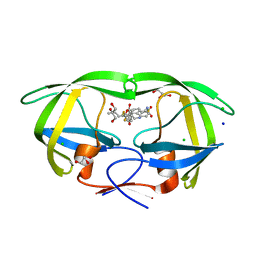 | | Wild-type HIV-1 protease in complex with tipranavir | | Descriptor: | CHLORIDE ION, GLYCEROL, HIV-1 protease, ... | | Authors: | Wong-Sam, A.E, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
5DGU
 
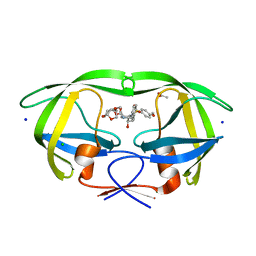 | | Crystal Structure of HIV-1 Protease Inhibitors Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand GRL-004-11A | | Descriptor: | (3R,3aR,4S,7aS)-3-methoxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
6E9A
 
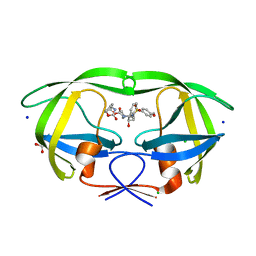 | | HIV-1 WILD TYPE PROTEASE WITH GRL-034-17A, (3aS, 5R, 6aR)-2-OXOHEXAHYD CYCLOPENTA[D]-5-OXAZOLYL URETHANE WITH A BICYCLIC OXAZOLIDINONE SCAFF AS THE P2 LIGAND | | Descriptor: | (3aS,5R,6aR)-2-oxohexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
6DJ1
 
 | | Wild-type HIV-1 protease in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
8F0F
 
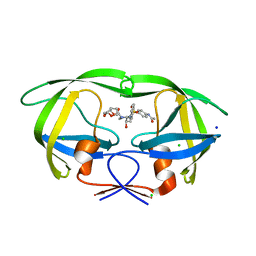 | | HIV-1 wild type protease with GRL-110-19A, a chloroacetamide derivative based on Darunavir as P2' group | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[4-(2-chloroacetamido)benzene-1-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.
Bioorg.Med.Chem.Lett., 83, 2023
|
|
6E7J
 
 | | HIV-1 wild type protease with GRL-042-17A, 3-phenylhexahydro-2h-cyclopenta[d]oxazol-2-one with a bicyclic oxazolidinone scaffold as the P2 ligand | | Descriptor: | (3aS,5R,6aR)-2-oxo-3-phenylhexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
6DJ7
 
 | | HIV-1 protease with mutation L76V in complex with GRL-5010 (gem-difluoro-bis-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
8ESX
 
 | | HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
8ESY
 
 | | D30N mutant HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
6DJ2
 
 | | HIV-1 protease with single mutation L76V in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
4U7V
 
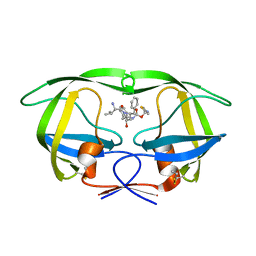 | | Structure of wild-type HIV protease in complex with degraded photosensitive inhibitor | | Descriptor: | BETA-MERCAPTOETHANOL, N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
4U1J
 
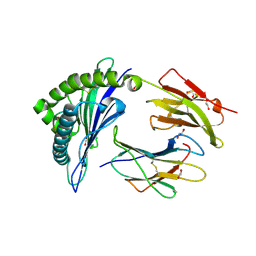 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
3QIO
 
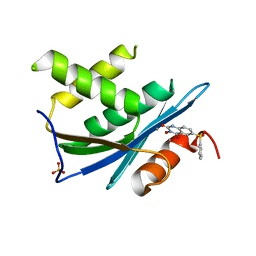 | | Crystal Structure of HIV-1 RNase H with engineered E. coli loop and N-hydroxy quinazolinedione inhibitor | | Descriptor: | 3-hydroxy-6-(phenylsulfonyl)quinazoline-2,4(1H,3H)-dione, Gag-Pol polyprotein,Ribonuclease HI,Gag-Pol polyprotein, MANGANESE (II) ION, ... | | Authors: | Lansdon, E.B, Liu, Q. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4011 Å) | | Cite: | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
6DIL
 
 | | HIV-1 protease with single mutation L76V in complex with tipranavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
4Q1Y
 
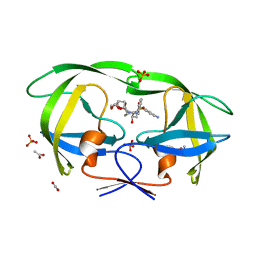 | | Mutations Outside the Active Site of HIV-1 Protease Alter Enzyme Structure and Dynamic Ensemble of the Active Site to Confer Drug Resistance | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Ragland, D.A, Nalam, M.N.L, Cao, H, Nalivaika, E.A, Cai, Y, Kurt-Yilmaz, N, Schiffer, C.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease.
J.Am.Chem.Soc., 136, 2014
|
|
6J1W
 
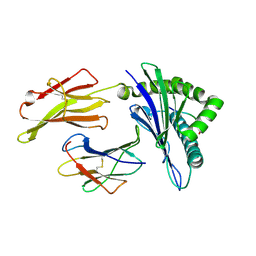 | | The structure of HLA-A*3001/RT313 | | Descriptor: | ALA-ILE-PHE-GLN-SER-SER-MET-THR-LYS, Beta-2-microglobulin, HLA-A*3001 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6PYL
 
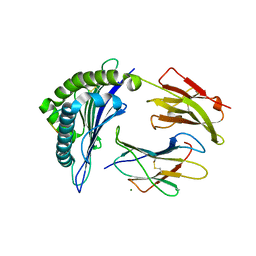 | | Crystal Structure of HLA-B*2703 in complex with KK10, an HIV peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27:03 alpha chain, ... | | Authors: | Gras, S. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Allelic association with ankylosing spondylitis fails to correlate with human leukocyte antigen B27 homodimer formation.
J.Biol.Chem., 294, 2019
|
|
4U1H
 
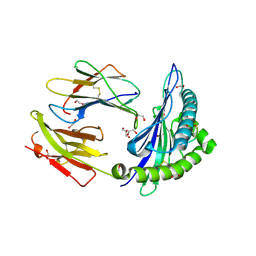 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
7R7V
 
 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
1EXQ
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5DGW
 
 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | Descriptor: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
7DPQ
 
 | | HIV-1 Protease D30N mutant | | Descriptor: | Protease | | Authors: | Bihani, S.C, Hosur, M.V. | | Deposit date: | 2020-12-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for reduced cleavage activity and drug resistance in D30N HIV-1 protease.
J.Biomol.Struct.Dyn., 2021
|
|
2NPH
 
 | | Crystal structure of HIV1 protease in situ product complex | | Descriptor: | PROTEASE RETROPEPSIN, pentapeptide fragment, tetrapeptide fragment | | Authors: | Hosur, M.V, Das, A, Prashar, V. | | Deposit date: | 2006-10-27 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HIV-1 protease in situ product complex and observation of a low-barrier hydrogen bond between catalytic aspartates
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
