1SLN
 
 | |
1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLQ
 
 | | Crystal structure of the trimeric state of the rhesus rotavirus VP4 membrane interaction domain, VP5CT | | Descriptor: | VP4 | | Authors: | Dormitzer, P.R, Nason, E.B, Prasad, B.V.V, Harrison, S.C. | | Deposit date: | 2004-03-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural rearrangements in the membrane penetration protein of a non-enveloped virus.
Nature, 430, 2004
|
|
1SLS
 
 | | IMMOBILE SLIPPED-LOOP STRUCTURE (SLS) OF DNA HOMODIMER IN SOLUTION, NMR, 9 STRUCTURES | | Descriptor: | OLIGODEOXYRIBONUCLEOTIDE | | Authors: | Ulyanov, N.B, Ivanov, V.I, Minyat, E.E, Khomyakova, E.B, Petrova, M.V, Lesiak, K, James, T.L. | | Deposit date: | 1997-09-30 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A pseudosquare knot structure of DNA in solution.
Biochemistry, 37, 1998
|
|
1SLT
 
 | |
1SLU
 
 | |
1SLV
 
 | |
1SLW
 
 | |
1SLX
 
 | |
1SLY
 
 | | COMPLEX OF THE 70-KDA SOLUBLE LYTIC TRANSGLYCOSYLASE WITH BULGECIN A | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, 70-KDA SOLUBLE LYTIC TRANSGLYCOSYLASE | | Authors: | Thunnissen, A.M.W.H, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1995-08-02 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism.
Biochemistry, 34, 1995
|
|
1SM1
 
 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH QUINUPRISTIN AND DALFOPRISTIN | | Descriptor: | 23S RIBOSOMAL RNA, 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Harms, J.M, Schluenzen, F, Fucini, P, Bartels, H, Yonath, A. | | Deposit date: | 2004-03-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Alterations at the Peptidyl Transferase Centre of the Ribosome Induced by the Synergistic Action of the Streptogramins Dalfopristin and Quinupristin.
Bmc Biol., 2, 2004
|
|
1SM2
 
 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
1SM3
 
 | | CRYSTAL STRUCTURE OF THE TUMOR SPECIFIC ANTIBODY SM3 COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDE EPITOPE, ... | | Authors: | Dokurno, P, Bates, P.A, Band, H.A, Stewart, L.M.D, Lally, J.M, Burchell, J.M, Taylor-Papadimitriou, J, Sternberg, M.J.E, Snary, D, Freemont, P.S. | | Deposit date: | 1997-12-23 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure at 1.95 A resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition.
J.Mol.Biol., 284, 1998
|
|
1SM4
 
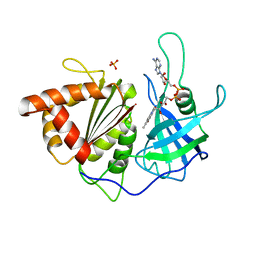 | | Crystal Structure Analysis of the Ferredoxin-NADP+ Reductase from Paprika | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, chloroplast ferredoxin-NADP+ oxidoreductase | | Authors: | Dorowski, A, Hofmann, A, Steegborn, C, Boicu, M, Huber, R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of paprika ferredoxin-NADP+ reductase. Implications for the electron transfer pathway.
J.Biol.Chem., 276, 2001
|
|
1SM5
 
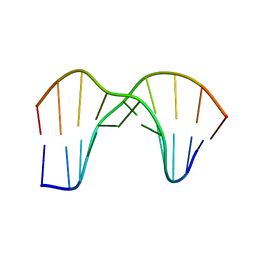 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2004-03-08 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SM7
 
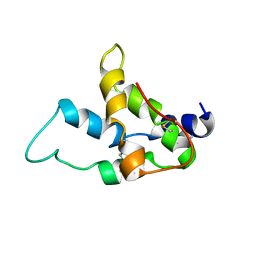 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
1SM8
 
 | | M. tuberculosis dUTPase complexed with chromium and dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHROMIUM ION, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SM9
 
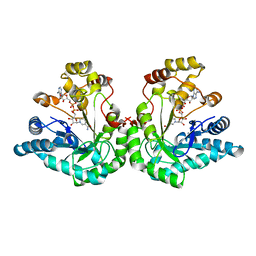 | | Crystal Structure Of An Engineered K274RN276D Double Mutant of Xylose Reductase From Candida Tenuis Optimized To Utilize NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Petschacher, B, Leitgeb, S, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography.
Biochem.J., 385, 2005
|
|
1SMA
 
 | | CRYSTAL STRUCTURE OF A MALTOGENIC AMYLASE | | Descriptor: | MALTOGENIC AMYLASE | | Authors: | Kim, J.S, Cha, S.S, Oh, B.H. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a maltogenic amylase provides insights into a catalytic versatility.
J.Biol.Chem., 274, 1999
|
|
1SMB
 
 | | Crystal Structure of Golgi-Associated PR-1 protein | | Descriptor: | 17kD fetal brain protein | | Authors: | Serrano, R.L, Kuhn, A, Hendricks, A, Helms, J.B, Sinning, I, Groves, M.R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-09-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism
J.Mol.Biol., 339, 2004
|
|
1SMC
 
 | | Mycobacterium tuberculosis dUTPase complexed with dUTP in the absence of metal ion. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SMD
 
 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1SME
 
 | | PLASMEPSIN II, A HEMOGLOBIN-DEGRADING ENZYME FROM PLASMODIUM FALCIPARUM, IN COMPLEX WITH PEPSTATIN A | | Descriptor: | PLASMEPSIN II, Pepstatin | | Authors: | Silva, A.M, Lee, A.Y, Gulnik, S.V, Goldberg, D.E, Erickson, J.W. | | Deposit date: | 1996-06-11 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SMF
 
 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
