8OXI
 
 | |
8OXH
 
 | |
8OXA
 
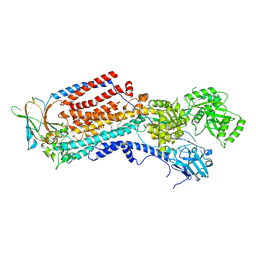 | | Cryo-EM structure of ATP8B1-CDC50A in E2-Pi conformation with occluded PS | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8OX9
 
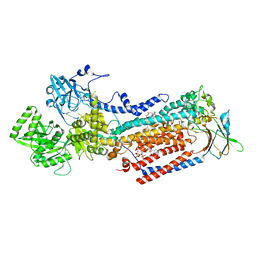 | | Cryo-EM structure of ATP8B1-CDC50A in E2P active conformation with bound PC | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8OX8
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited "open" conformation | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8OX7
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited "closed" conformation | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8OWZ
 
 | | Crystal structure of human Sirt2 in complex with a triazole-based SirReal | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[5-[[3-[[1-(2-methoxyethyl)-1,2,3-triazol-4-yl]methoxy]phenyl]methyl]-1,3-thiazol-2-yl]ethanamide, ... | | Authors: | Friedrich, F, Zhang, L, Schiedel, M, Einsle, O, Jung, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of First-in-Class Dual Sirt2/HDAC6 Inhibitors as Molecular Tools for Dual Inhibition of Tubulin Deacetylation.
J.Med.Chem., 66, 2023
|
|
8OWX
 
 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 2024
|
|
8OWW
 
 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | Authors: | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWV
 
 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWR
 
 | |
8OWQ
 
 | |
8OWP
 
 | |
8OWO
 
 | | SMYD3 in complex with fragment FL01507 | | Descriptor: | 3-oxidanylbenzenecarbonitrile, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening.
Rsc Med Chem, 15, 2024
|
|
8OWM
 
 | | Crystal structure of glutamate dehydrogenase 2 from Arabidopsis thaliana binding Ca, NAD and 2,2-dihydroxyglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(oxidanyl)pentanedioic acid, CALCIUM ION, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional studies of Arabidopsis thaliana glutamate dehydrogenase isoform 2 demonstrate enzyme dynamics and identify its calcium binding site.
Plant Physiol Biochem., 201, 2023
|
|
8OWI
 
 | |
8OWB
 
 | |
8OW5
 
 | |
8OW4
 
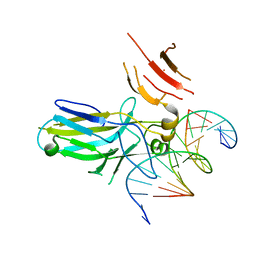 | | 2.75 angstrom crystal structure of human NFAT1 with bound DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*TP*TP*CP*CP*AP*GP*C)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*GP*GP*AP*AP*AP*AP*AP*TP*AP*G)-3'), Nuclear factor of activated T-cells, ... | | Authors: | Lopez-Sagaseta, J, Erausquin, E, Hernandez-Morales, S, Urdiciain, A, Lasarte, J.J, Lozano, T. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 angstrom crystal structure of human NFAT1 with bound DNA
Not published
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | Descriptor: | Green fluorescent protein | | Authors: | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
8OVQ
 
 | |
8OVE
 
 | |
8OVB
 
 | |
8OVA
 
 | |
8OV9
 
 | |
