1U5S
 
 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | Descriptor: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | Authors: | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | Deposit date: | 2004-07-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
1U5T
 
 | | Structure of the ESCRT-II endosomal trafficking complex | | Descriptor: | Defective in vacuolar protein sorting; Vps36p, Hypothetical 23.6 kDa protein in YUH1-URA8 intergenic region, appears to be functionally related to SNF7; Snf8p | | Authors: | Hierro, A, Sun, J, Rusnak, A.S, Kim, J, Prag, G, Emr, S.D, Hurley, J.H. | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of ESCRT-II endosomal trafficking complex
Nature, 431, 2004
|
|
1U5U
 
 | |
1U5V
 
 | | Structure of CitE complexed with triphosphate group of ATP form Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, citE | | Authors: | Goulding, C.W, Lerkin, T, Kim, C.Y, Segelke, B, Terwilliger, T, Eisenberg, E, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis citrate lyase beta subunit and its unusual triphosphate binding site
To be Published
|
|
1U5W
 
 | |
1U5X
 
 | | Crystal structure of murine APRIL at pH 5.0 | | Descriptor: | Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U5Y
 
 | | Crystal structure of murine APRIL, pH 8.0 | | Descriptor: | Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U5Z
 
 | | The Crystal structure of murine APRIL, pH 8.5 | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U60
 
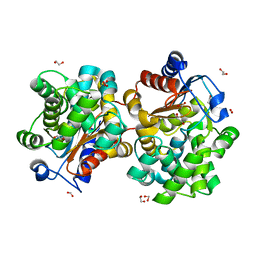 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
1U61
 
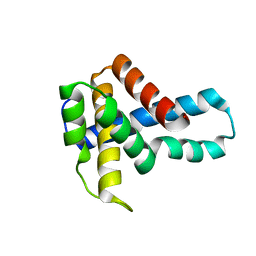 | | Crystal Structure of Putative Ribonuclease III from Bacillus cereus | | Descriptor: | hypothetical protein | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein from Bacillus cereus
To be Published
|
|
1U62
 
 | |
1U63
 
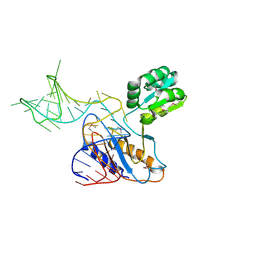 | | THE STRUCTURE OF A RIBOSOMAL PROTEIN L1-mRNA COMPLEX | | Descriptor: | 49 NT FRAGMENT OF MRNA FOR L1, 50S ribosomal protein L1P | | Authors: | Nevskaya, N, Tishchenko, S, Gabdoulkhakov, A, Nikonova, E, Nikonov, O, Nikulin, A, Garber, M, Nikonov, S, Piendl, W. | | Deposit date: | 2004-07-29 | | Release date: | 2005-04-12 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA.
Nucleic Acids Res., 33, 2005
|
|
1U64
 
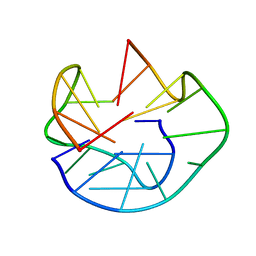 | | The Solution Structure of d(G3T4G4)2 | | Descriptor: | 5'-D(*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3' | | Authors: | Sket, P, Crnugelj, M, Plavec, J. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions.
Bioorg.Med.Chem., 12, 2004
|
|
1U65
 
 | | Ache W. CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Harel, M, Hyatt, J.L, Brumshtein, B, Morton, C.L, Wadkins, R.W, Silman, I, Sussman, J.L, Potter, P.M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-07-29 | | Release date: | 2005-07-19 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action
Mol.Pharmacol., 67, 2005
|
|
1U67
 
 | | Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Harman, C.A, Rieke, C.J, Garavito, R.M, Smith, W.L. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of arachidonic Acid bound to a mutant of prostaglandin endoperoxide h synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic Acid.
J.Biol.Chem., 279, 2004
|
|
1U68
 
 | | DHNA 7,8 DIHYDRONEOPTERIN COMPLEX | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization.
J.MED.CHEM., 47, 2004
|
|
1U69
 
 | | Crystal Structure of PA2721 Protein of Unknown Function from Pseudomonas aeruginosa PAO1 | | Descriptor: | hypothetical protein | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-21 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6 A crystal structure of a PA2721 protein from pseudomonas aeruginosa--a potential drug-resistance protein.
Proteins, 63, 2006
|
|
1U6A
 
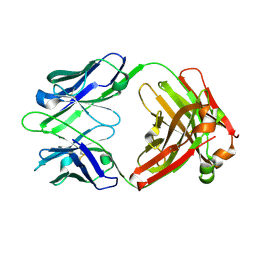 | | Crystal Structure of the Broadly Neutralizing Anti-HIV Fab F105 | | Descriptor: | F105 HEAVY CHAIN, F105 LIGHT CHAIN | | Authors: | Wilkinson, R.A, Piscitelli, C, Teintze, M, Lawrence, C.M. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120.
J.Virol., 79, 2005
|
|
1U6B
 
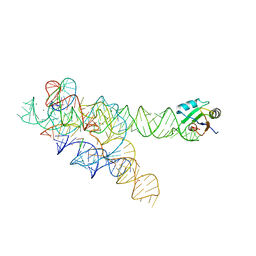 | | CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS | | Descriptor: | 197-MER, 5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*AP*GP*AP*CP*GP *GP*CP*C)-3', 5'-R(*CP*AP*(5MU))-3', ... | | Authors: | Adams, P.L, Stahley, M.R, Kosek, A.B, Wang, J, Strobel, S.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Self-Splicing Group I Intron with Both Exons.
Nature, 430, 2004
|
|
1U6C
 
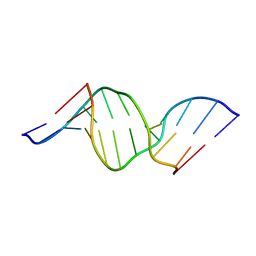 | | The NMR-derived solution structure of the (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide | | Descriptor: | (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide | | Authors: | Scholdberg, T.A, Nechev, L.N, Merritt, W.K, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a site specific major groove (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide.
Chem.Res.Toxicol., 17, 2004
|
|
1U6D
 
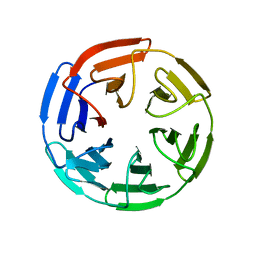 | |
1U6E
 
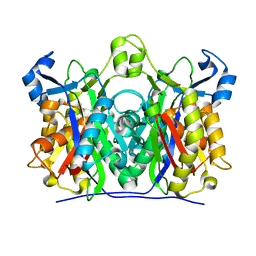 | | 1.85 Angstrom Crystal Structure of the C112A Mutant of Mycobacterium Tuberculosis Beta-Ketoacyl-Acyl Carrier Protein Synthase III (FabH) | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III, CHLORIDE ION | | Authors: | Mussayev, F, Sachedeva, S, Scarsdale, J.N, Reynolds, K.A, Wright, H.T. | | Deposit date: | 2004-07-29 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A.
J.Mol.Biol., 346, 2005
|
|
1U6F
 
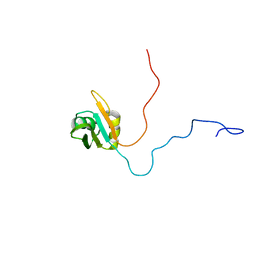 | | NMR solution structure of TcUBP1, a single RBD-unit from Trypanosoma cruzi | | Descriptor: | RNA-binding protein UBP1 | | Authors: | Volpon, L, D'orso, I, Frasch, A, Gehring, K. | | Deposit date: | 2004-07-29 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Study of TcUBP1, a Single RRM Domain Protein from Trypanosoma cruzi: Contribution of a beta Hairpin to RNA Binding
Biochemistry, 44, 2005
|
|
1U6G
 
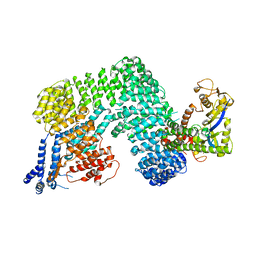 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
1U6H
 
 | | Vinculin head (0-258) in complex with the talin vinculin binding site 2 (849-879) | | Descriptor: | Talin, Vinculin | | Authors: | Fillingham, I, Gingras, A.R, Papagrigoriou, E, Patel, B, Emsley, J, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2004-07-30 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head.
Structure, 13, 2005
|
|
