4QQU
 
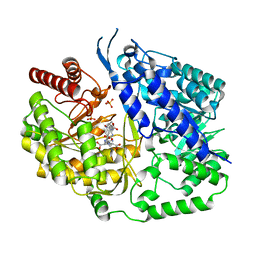 | | Crystal structure of the cobalamin-independent methionine synthase enzyme in a closed conformation | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, N-[4-({[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-gamma-glutamyl-L-glutamic acid, ... | | Authors: | Ubhi, D.K, Robertus, J.D. | | Deposit date: | 2014-06-29 | | Release date: | 2015-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The cobalamin-independent methionine synthase enzyme captured in a substrate-induced closed conformation.
J.Mol.Biol., 427, 2015
|
|
7BES
 
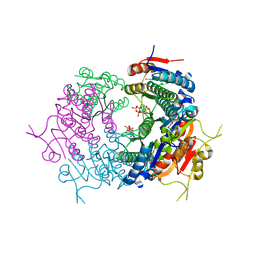 | | CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Bous, J, Trapani, S, Walter, P, Bron, P, Munier-Lehmann, H. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
4R7G
 
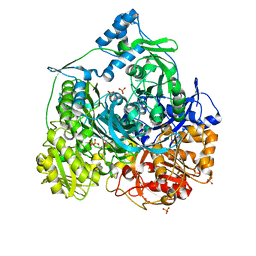 | | Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Tanwar, A.S, Sindhikara, D.J, Hirata, F, Anand, R. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment.
Acs Chem.Biol., 10, 2015
|
|
3EWX
 
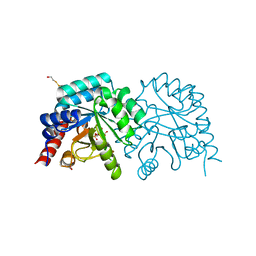 | | K314A mutant of human orotidyl-5'-monophosphate decarboxylase in complex with 6-azido-UMP, degraded to BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine-5'-phosphate decarboxylase | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
3FJ6
 
 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 2 | | Descriptor: | (2Z)-2-cyano-N-(2,2'-dichlorobiphenyl-4-yl)-3-hydroxybut-2-enamide, ACETATE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Heikkila, T. | | Deposit date: | 2008-12-14 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
3G1D
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4R52
 
 | | 1.5 angstrom crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Geng, J, Gumpper, R.H, Huo, L, Liu, A. | | Deposit date: | 2014-08-20 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | 1.5 angstrom crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To be Published
|
|
4IZO
 
 | |
4YCD
 
 | |
4R5M
 
 | | Crystal structure of Vc-Aspartate beta-semialdehyde-dehydrogenase with NADP and 4-Nitro-2-Phosphono-Benzoic acid | | Descriptor: | 4-nitro-2-phosphonobenzoic acid, Aspartate-semialdehyde dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pavlovsky, A.G, Thangavelu, B, Bhansali, P, Viola, R.E. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3G3M
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-iodo-UMP | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Poduch, E, Kotra, L.P, Pai, E.F. | | Deposit date: | 2009-02-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
3G0U
 
 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 4 | | Descriptor: | (2Z)-N-(3-chloro-2'-methoxybiphenyl-4-yl)-2-cyano-3-hydroxybut-2-enamide, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Davis, M, Heikkila, T, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2009-01-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
3N0V
 
 | |
3G0X
 
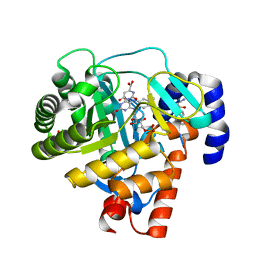 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 5 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enamide, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Davies, M, Heikkila, T, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
3G18
 
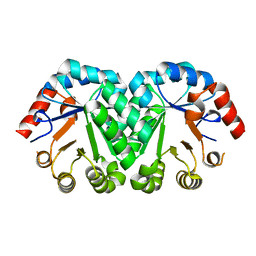 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4RX0
 
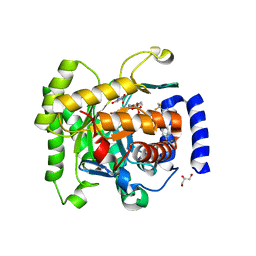 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
3G3D
 
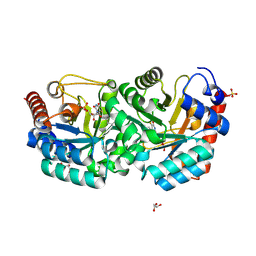 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-azido-UMP | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, Y, Tang, H.L, Bello, A, Poduch, E, Kotra, L, Pai, E. | | Deposit date: | 2009-02-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
4YCC
 
 | |
7UXH
 
 | | cryo-EM structure of the mTORC1-TFEB-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Cui, Z, Hurley, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex.
Nature, 614, 2023
|
|
7UXC
 
 | |
3G1A
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4Z7J
 
 | | Structure of Acetobacter aceti PurE-S57A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 5-O-phosphono-beta-D-ribofuranose, ... | | Authors: | Kappock, T.J, Sullivan, K.L. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Mutation of the conserved serine in two PurE classes
To Be Published
|
|
4ZJY
 
 | |
4ZMB
 
 | |
4WZC
 
 | |
