1E2A
 
 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | Descriptor: | ENZYME IIA, MAGNESIUM ION | | Authors: | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
1CLF
 
 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
2L8S
 
 | |
6LY5
 
 | | Organization and energy transfer in a huge diatom PSI-FCPI supercomplex | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Xiong, P, Caizhe, X. | | Deposit date: | 2020-02-13 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex.
Nat Commun, 11, 2020
|
|
2MTG
 
 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2DG9
 
 | |
2DG4
 
 | | FK506-binding protein mutant WF59 complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2DG3
 
 | | Wildtype FK506-binding protein complexed with Rapamycin | | Descriptor: | FK506-binding protein 1A, GLYCEROL, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Buckle, A.M. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Energetic and structural analysis of the role of tryptophan 59 in FKBP12
Biochemistry, 42, 2003
|
|
2NAV
 
 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2016-08-17 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
2MFN
 
 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
1YUF
 
 | |
1YUG
 
 | |
7R09
 
 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ and Cyclohexylamine | | Descriptor: | Amine Dehydrogenase, CYCLOHEXYLAMMONIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
5B34
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5C2M
 
 | | The de novo evolutionary emergence of a symmetrical protein is shaped by folding constraints | | Descriptor: | Predicted protein | | Authors: | Smock, R.G, Yadid, I, Dym, O, Clarke, J, Tawfik, D.S. | | Deposit date: | 2015-06-16 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De Novo Evolutionary Emergence of a Symmetrical Protein Is Shaped by Folding Constraints.
Cell, 164, 2016
|
|
1BPJ
 
 | | THYMIDYLATE SYNTHASE R178T, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
5C2N
 
 | | The de novo evolutionary emergence of a symmetrical protein is shaped by folding constraints | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta propeller | | Authors: | Smock, R.G, Yadid, I, Dym, O, Clarke, J, Tawfik, D.S. | | Deposit date: | 2015-06-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De Novo Evolutionary Emergence of a Symmetrical Protein Is Shaped by Folding Constraints.
Cell, 164, 2016
|
|
1BO8
 
 | | THYMIDYLATE SYNTHASE R178T MUTANT | | Descriptor: | POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE), URIDINE-5'-MONOPHOSPHATE | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-10 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BP0
 
 | | THYMIDYLATE SYNTHASE R23I MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BO7
 
 | | THYMIDYLATE SYNTHASE R179T MUTANT | | Descriptor: | PROTEIN (THYMIDYLATE SYNTHASE), URIDINE-5'-MONOPHOSPHATE | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-10 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BP6
 
 | | THYMIDYLATE SYNTHASE R23I, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
6OF1
 
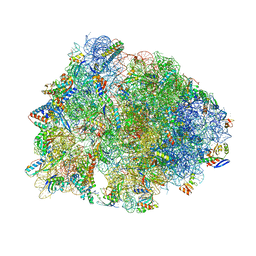 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with dirithromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khabibullina, N.F, Tereshchenkov, A.G, Komarova, E.S, Syroegin, E.A, Shiriaev, D.I, Paleskava, A, Kartsev, V.G, Bogdanov, A.A, Konevega, A.L, Dontsova, O.A, Sergiev, P.V, Osterman, I.A, Polikanov, Y.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Dirithromycin Bound to the Bacterial Ribosome Suggests New Ways for Rational Improvement of Macrolides.
Antimicrob.Agents Chemother., 63, 2019
|
|
7LUX
 
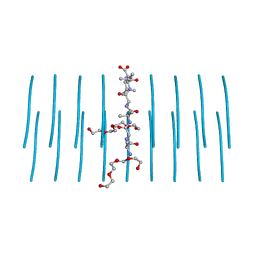 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7LUZ
 
 | |
