5M5K
 
 | | S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine and cordycepin | | Descriptor: | 3'-DEOXYADENOSINE, ACETATE ION, ADENOSINE, ... | | Authors: | Manszewski, T, Mueller-Dieckamann, J, Jaskolski, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
8QNO
 
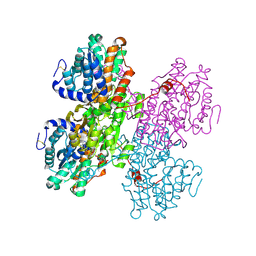 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase treated at 368 K from Pyrococcus furiosus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Koeppl, L.H, Popadic, D, Andexer, J.N. | | Deposit date: | 2023-09-27 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
8OLR
 
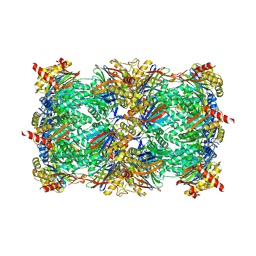 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLL
 
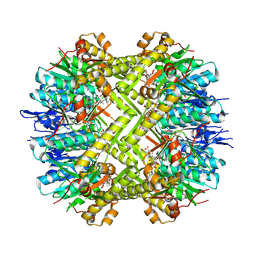 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1C7N
 
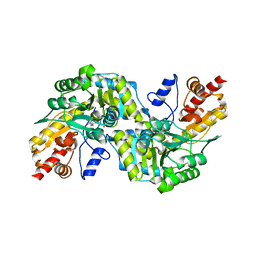 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE COFACTOR | | Descriptor: | CYSTALYSIN, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
6G7F
 
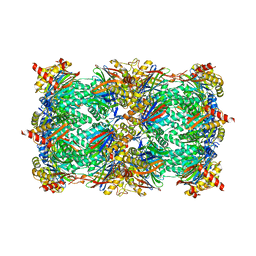 | | Yeast 20S proteasome in complex with Cystargolide B | | Descriptor: | CHLORIDE ION, Cystargolide B- bound form, MAGNESIUM ION, ... | | Authors: | Groll, M, Tello-Aburto, R. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
8WZ6
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep | | Descriptor: | Adenosylhomocysteinase, DZNep, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WZ8
 
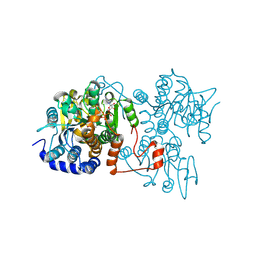 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD | | Descriptor: | Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WWG
 
 | |
4QJZ
 
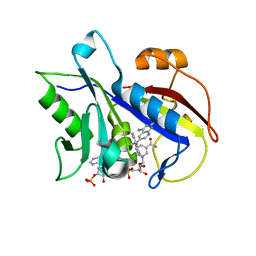 | | Pneumocystis carinii dihydrofolate reductase ternary complex with NADPH and the inhibitor 24, (N~6~-METHYL-N~6~-(NAPHTHALEN-1-YL)PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(naphthalen-1-yl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
7CR6
 
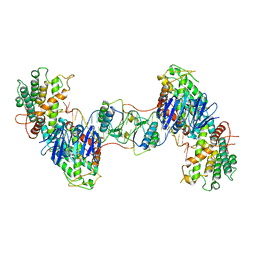 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CR8
 
 | | Synechocystis Cas1-Cas2-prespacerL complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
2N8C
 
 | |
1W04
 
 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Glycine-Fe-NO Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-GLYCINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
1W06
 
 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Alanine-Fe NO Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-ALANINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
4OJT
 
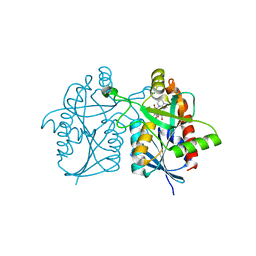 | |
4F3K
 
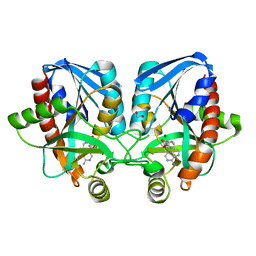 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with homocysteine-DADMe-Immucillin-A | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, {(3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-hydroxypyrrolidin-3-yl}-L-methionine | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with homocysteine-DADMe-Immucillin-A
Structure, 21, 2013
|
|
2N8B
 
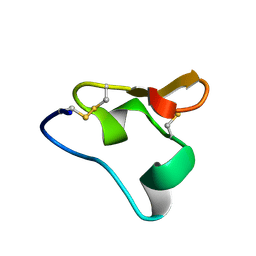 | |
1IM8
 
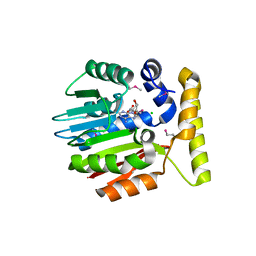 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
4MJC
 
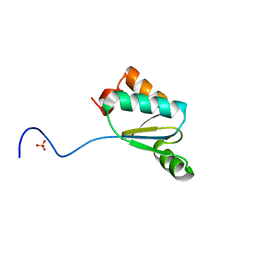 | | Synechocystis sp. PCC 6803 glutaredoxin A - P84R | | Descriptor: | Probable glutaredoxin ssr2061, SULFATE ION | | Authors: | Sutton, R.B, Knaff, D.B. | | Deposit date: | 2013-09-03 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Utility of Synechocystis sp. PCC 6803 glutaredoxin A as a platform to study high-resolution mutagenesis of proteins.
Front Plant Sci, 4, 2013
|
|
4MJB
 
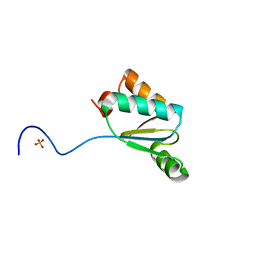 | | Synechocystis sp. PCC 6803 glutaredoxin A-A79S | | Descriptor: | Probable glutaredoxin ssr2061, SULFATE ION | | Authors: | Sutton, R.B, Knaff, D.B. | | Deposit date: | 2013-09-03 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Utility of Synechocystis sp. PCC 6803 glutaredoxin A as a platform to study high-resolution mutagenesis of proteins.
Front Plant Sci, 4, 2013
|
|
4A82
 
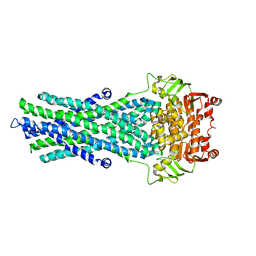 | | Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966. | | Descriptor: | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR | | Authors: | Rosenberg, M.F, ORyan, L.P, Hughes, G, Zhao, Z, Aleksandrov, L.A, Riordan, J.R, Ford, R.C. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | The Cystic Fibrosis Transmembrane Conductance Regulator (Cftr):3D Structure and Localisation of a Channel Gate.
J.Biol.Chem., 286, 2011
|
|
4MJE
 
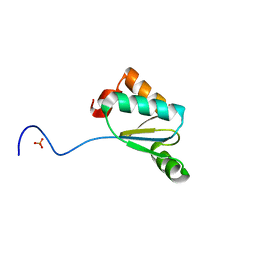 | | Synechocystis sp. PCC 6803 glutaredoxin A-R27L | | Descriptor: | Probable glutaredoxin ssr2061, SULFATE ION | | Authors: | Sutton, R.B, Knaff, D.B. | | Deposit date: | 2013-09-03 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Utility of Synechocystis sp. PCC 6803 glutaredoxin A as a platform to study high-resolution mutagenesis of proteins.
Front Plant Sci, 4, 2013
|
|
4MJA
 
 | | Synechocystis sp. PCC 6803 glutaredoxin A-A75I | | Descriptor: | Probable glutaredoxin ssr2061 | | Authors: | Sutton, R.B, Knaff, D.B. | | Deposit date: | 2013-09-03 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Utility of Synechocystis sp. PCC 6803 glutaredoxin A as a platform to study high-resolution mutagenesis of proteins.
Front Plant Sci, 4, 2013
|
|
2MHH
 
 | | Solution structure of a EF-hand domain from sea urchin polycystin-2 | | Descriptor: | Polycystic kidney disease protein 2 | | Authors: | Kuo, I.Y, Keeler, C, Corbin, R, Celic, A, Petri, E.T, Hodsdon, M.E, Ehrlich, B.E. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The number and location of EF hand motifs dictates the calcium dependence of polycystin-2 function.
Faseb J., 28, 2014
|
|
