8KAI
 
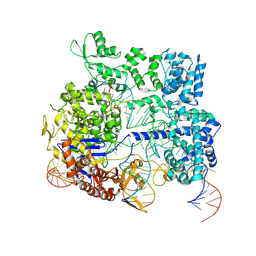 | | Crystal structure of SpyCas9-crRNA-tracrRNA complex bound to 17nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (25-MER), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 43, 2025
|
|
5NFV
 
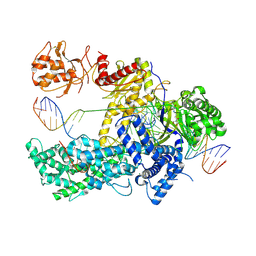 | | Crystal structure of catalytically inactive FnCas12 mutant bound to an R-loop structure containing a pre-crRNA mimic and full-length DNA target | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CRISPR-associated endonuclease Cpf1, DNA non-target strand, ... | | Authors: | Swarts, D.C, van der Oost, J, Jinek, M. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a.
Mol. Cell, 66, 2017
|
|
7VG2
 
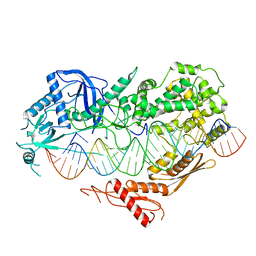 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 40-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a forward strand (5'-phosphorylation), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7ROU
 
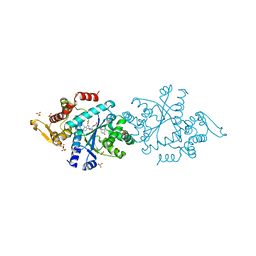 | | Structure of human tyrosyl tRNA synthetase in complex with ML901-Tyr | | Descriptor: | SULFATE ION, Tyrosine--tRNA ligase, cytoplasmic, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy.
Science, 376, 2022
|
|
7OS0
 
 | |
5GXH
 
 | | The structure of the Gemin5 WD40 domain with AAUUUUUG | | Descriptor: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*G)-3'), ... | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
6Z2B
 
 | |
5GXI
 
 | | Structure of the Gemin5 WD40 domain in complex with AAUUUUUGAG | | Descriptor: | Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*GP*AP*G)-3'), UNKNOWN ATOM OR ION | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
4PQU
 
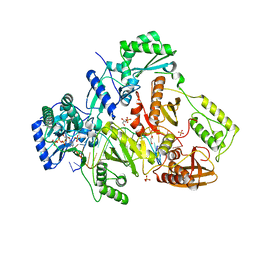 | | Crystal structure of HIV-1 Reverse Transcriptase in complex with RNA/DNA and dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', ... | | Authors: | Das, K, Bandwar, R.P, Arnold, E. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
6GPG
 
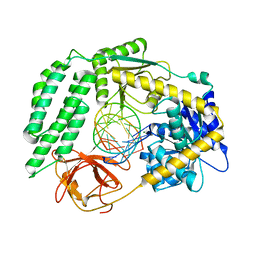 | | Structure of the RIG-I Singleton-Merten syndrome variant C268F | | Descriptor: | MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*CP*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*CP*G)-3'), ... | | Authors: | Laessig, C, Lammens, K, Hopfner, K.-P. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Unified mechanisms for self-RNA recognition by RIG-I Singleton-Merten syndrome variants.
Elife, 7, 2018
|
|
5UX0
 
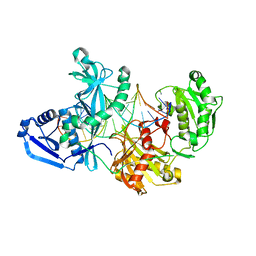 | |
7Y9Y
 
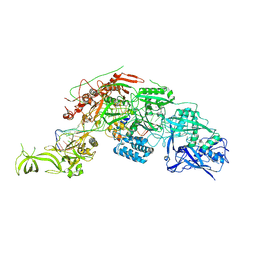 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y82
 
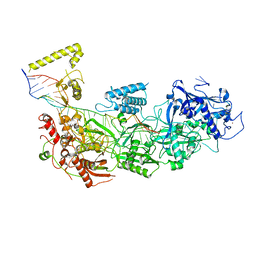 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to self RNA target | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, Self RNA target, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y83
 
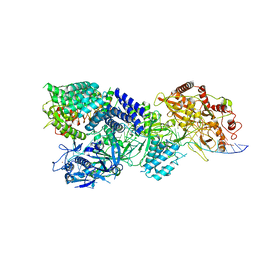 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to non-self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y81
 
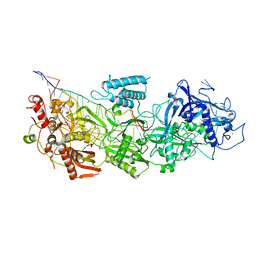 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to non-self RNA target | | Descriptor: | MAGNESIUM ION, Non-self RNA target, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y85
 
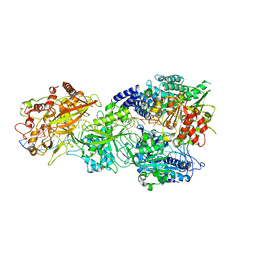 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
8Z1G
 
 | | Cryo-EM structure of human ELAC2-pre-tRNA | | Descriptor: | Homo sapiens mitochondrion pre-tRNA-Tyr, PHOSPHATE ION, ZINC ION, ... | | Authors: | Liu, Z.M, Xue, C.Y. | | Deposit date: | 2024-04-11 | | Release date: | 2024-12-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into human ELAC2 as a tRNA 3' processing enzyme.
Nucleic Acids Res., 52, 2024
|
|
8WUT
 
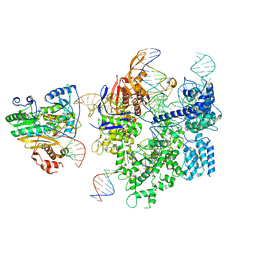 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
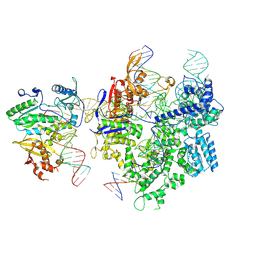 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
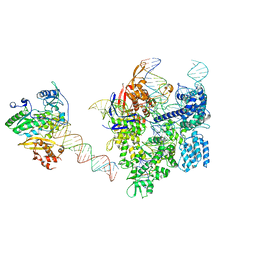 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
7Y9X
 
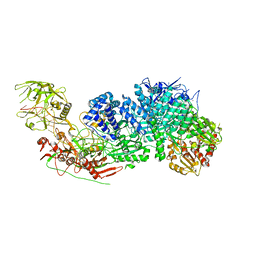 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7BY6
 
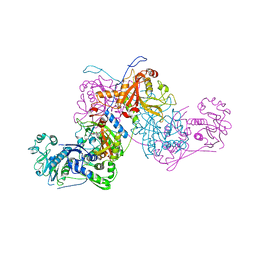 | | Plasmodium vivax cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD1389 | | Descriptor: | (3S,4R,8R,9R,10S)-N-(4-cyclopropyloxyphenyl)-10-(methoxymethyl)-3,4-bis(oxidanyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Malhotra, N, Manmohan, S, Harlos, K, Melillo, B, Schreiber, S.L, Manickam, Y, Sharma, S. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural basis of malaria parasite phenylalanine tRNA-synthetase inhibition by bicyclic azetidines.
Nat Commun, 12, 2021
|
|
3A2K
 
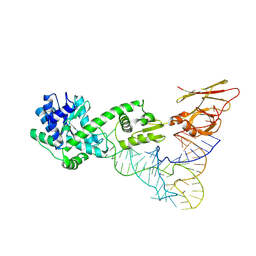 | | Crystal structure of TilS complexed with tRNA | | Descriptor: | bacterial tRNA, tRNA(Ile)-lysidine synthase | | Authors: | Nakanishi, K, Bonnefond, L, Ishitani, R, Nureki, O. | | Deposit date: | 2009-05-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
Nature, 461, 2009
|
|
2F4S
 
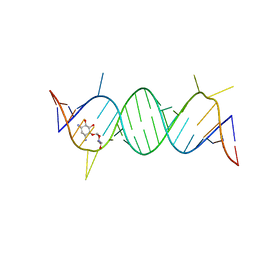 | | A-site RNA in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
