6VO6
 
 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
9NA4
 
 | | IRAK4 in Complex with Compound 18 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl {[(1S,3s)-3-{[(2P)-2-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-5-{[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]carbamoyl}pyridin-4-yl]amino}cyclobutyl]methyl}carbamate | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
7ZY5
 
 | | Crystal structure of compound 2 bound to CK2alpha | | Descriptor: | 1-NAPHTHOL, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
9RDI
 
 | | Crystal Structure of Flap Endonuclease FEN1 with Compound 5 | | Descriptor: | 2-methyl-9-oxidanyl-6-(phenylmethyl)-3,4-dihydropyrazino[1,2-c]pyrimidine-1,8-dione, 6-tungstotellurate(VI), Flap endonuclease 1, ... | | Authors: | Toste Rego, A, Pica, A, Cornaciu, I, Burgdorf, L, Mann, S.E. | | Deposit date: | 2025-06-02 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Fragment-Based Discovery and Structure-Led Optimization of MSC778, the First Potent, Selective, and Orally Bioavailable FEN1 Inhibitor.
J.Med.Chem., 2025
|
|
6NY7
 
 | | Crystal Structure of NDM-1 D199N with Compound 16 | | Descriptor: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5,7-dibromo-2-oxo-1,2-dihydroquinolin-4-yl)methyl]phosphonic acid | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-02-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
9Q8H
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative BDH 34019023 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Turk, D, Oberthuer, D, Kiene, A. | | Deposit date: | 2025-02-24 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
8TRS
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8OMM
 
 | | Coproporphyrin III - LmCpfC complex soaked 3min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, Coproporphyrin III ferrochelatase, FE (II) ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
6O3R
 
 | | Crystal Structure of NDM-1 D199N with Compound 7 | | Descriptor: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5-methoxy-7-methyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
9OWA
 
 | | Human WRN helicase in complex with allosteric ligand Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, CHLORIDE ION, ... | | Authors: | Palte, R.L, Koglin, M, Maskos, K, Tauchert, M.J. | | Deposit date: | 2025-06-02 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-throughput evaluation of novel WRN inhibitors.
Slas Discov, 35, 2025
|
|
8F1Y
 
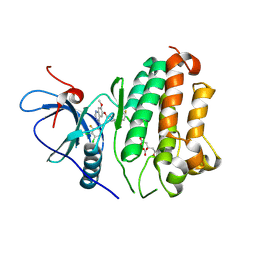 | | EGFR kinase in complex with poziotinib | | Descriptor: | 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, CITRIC ACID, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F1W
 
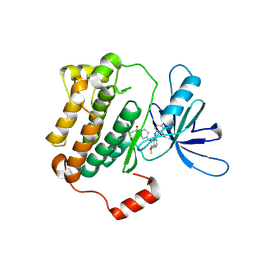 | | EGFR(T790M/V948R) kinase in complex with poziotinib | | Descriptor: | 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8CNC
 
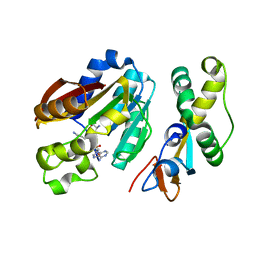 | | Structure of compound 1 bound KMT9 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein | | Authors: | Sheng, W. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of compound 1 bound KMT9
To Be Published
|
|
8CNO
 
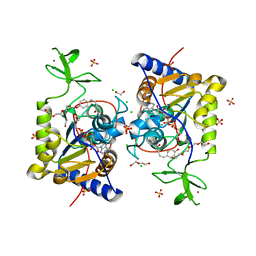 | |
7MAG
 
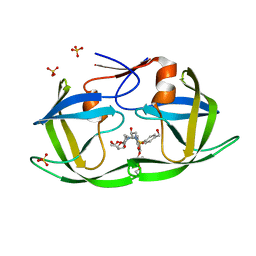 | | HIV-1 Protease (I84V) in Complex with PU3 (LR3-69) | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
8TXY
 
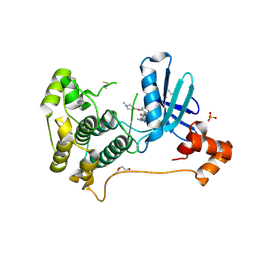 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7MAK
 
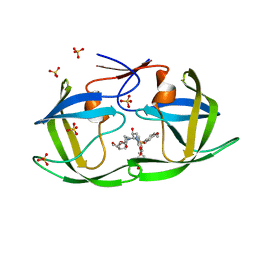 | | HIV-1 Protease (I84V) in Complex with PU7 (LR3-67) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{(2-ethylbutyl)[(4-methoxyphenyl)sulfonyl]amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
4R2S
 
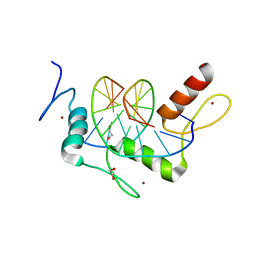 | | Wilms Tumor Protein (WT1) Q369P zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
7MAJ
 
 | | HIV-1 Protease (I84V) in Complex with PU6 (LR3-66) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
6VNQ
 
 | |
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
1DUW
 
 | | STRUCTURE OF NONAHEME CYTOCHROME C | | Descriptor: | GLYCEROL, HEME C, NONAHEME CYTOCHROME C | | Authors: | Umhau, S, Fritz, G, Diederichs, K, Breed, J, Kroneck, P.M, Welte, W. | | Deposit date: | 2000-01-19 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Three-dimensional structure of the nonaheme cytochrome c from Desulfovibrio desulfuricans Essex in the Fe(III) state at 1.89 A resolution.
Biochemistry, 40, 2001
|
|
7PIL
 
 | | Cryo-EM structure of the Rhodobacter sphaeroides RC-LH1-PufXY monomer complex at 2.5 A | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the monomeric Rhodobacter sphaeroides RC-LH1 core complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
6VVQ
 
 | |
8ORI
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (orthorhombic) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
