2AT2
 
 | |
1RHM
 
 | |
2BC2
 
 | |
2AZO
 
 | | DNA MISMATCH REPAIR PROTEIN MUTH FROM E. COLI | | Descriptor: | MUTH | | Authors: | Yang, W. | | Deposit date: | 1997-11-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases.
EMBO J., 17, 1998
|
|
2ALP
 
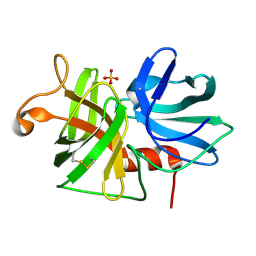 | | REFINED STRUCTURE OF ALPHA-LYTIC PROTEASE AT 1.7 ANGSTROMS RESOLUTION. ANALYSIS OF HYDROGEN BONDING AND SOLVENT STRUCTURE | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Fujinaga, M, Delbaere, L.T.J, Brayer, G.D, James, M.N.G. | | Deposit date: | 1985-03-07 | | Release date: | 1985-07-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure.
J.Mol.Biol., 184, 1985
|
|
2AT9
 
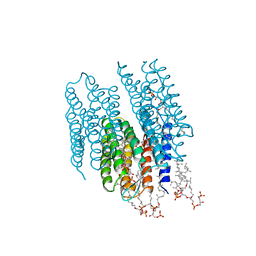 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | 3-[[3-METHYLPHOSPHONO-GLYCEROLYL]PHOSPHONYL]-[1,2-DI[2,6,10,14-TETRAMETHYL-HEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, RETINAL | | Authors: | Mitsuoka, K, Hirai, T, Murata, K, Miyazawa, A, Kidera, A, Kimura, Y, Fujiyoshi, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution.
J.Mol.Biol., 286, 1999
|
|
1RMH
 
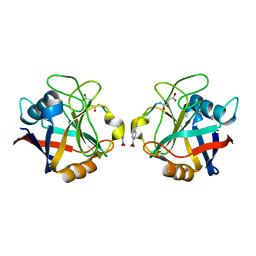 | | RECOMBINANT CYCLOPHILIN A FROM HUMAN T CELL | | Descriptor: | AAPF PEPTIDE SUBSTRATE, CYCLOPHILIN A | | Authors: | Zhao, Y, Ke, H. | | Deposit date: | 1995-07-31 | | Release date: | 1996-10-14 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure implies that cyclophilin predominantly catalyzes the trans to cis isomerization.
Biochemistry, 35, 1996
|
|
2BBQ
 
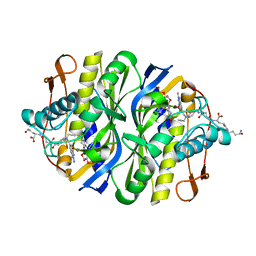 | | STRUCTURAL BASIS FOR RECOGNITION OF POLYGLUTAMYL FOLATES BY THYMIDYLATE SYNTHASE | | Descriptor: | 10-PARPARGYL-5,8-DIDEAZAFOLATE-4-GLUTAMIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Kamb, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1992-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of polyglutamyl folates by thymidylate synthase.
Biochemistry, 31, 1992
|
|
1RN7
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1RNR
 
 | |
2BEB
 
 | | X-ray structure of Asn to Thr mutant of Winged Bean Chymotrypsin inhibitor | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2BE5
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
1RVJ
 
 | | PHOTOSYNTHETIC REACTION CENTER DOUBLE MUTANT FROM RHODOBACTER SPHAEROIDES WITH ASP L213 REPLACED WITH ASN AND ARG H177 REPLACED WITH HIS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Xu, Q, Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2003-12-14 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | X-Ray Structure Determination of Three Mutants of the Bacterial Photosynthetic Reaction
Centers from Rb. sphaeroides; Altered Proton Transfer Pathways.
Structure, 12, 2004
|
|
1Z94
 
 | | X-Ray Crystal Structure of Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR12. | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A, Vorobiev, S.M, Yong, W, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CVR12.
To be Published
|
|
1RWE
 
 | | Enhancing the activity of insulin at receptor edge: crystal structure and photo-cross-linking of A8 analogues | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z, Xu, B, Chu, Y.C, Li, B, Nakagawa, S.H, Qu, Y, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues.
Biochemistry, 43, 2004
|
|
1R2F
 
 | |
1R2Y
 
 | | MutM (Fpg) bound to 8-oxoguanine (oxoG) containing DNA | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA Lesion Recognition by the Bacterial Repair Enzyme MutM.
J.Biol.Chem., 278, 2003
|
|
1R35
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER, TETRAHYDROBIOPTERIN AND 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE | | Descriptor: | 4R-FLUORO-N6-ETHANIMIDOYL-L-LYSINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Shieh, H.S, Stevens, A.M, Stallings, W.C. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4-Fluorinated L-lysine analogs as selective i-NOS inhibitors: methodology for introducing fluorine into the lysine side chain.
Org.Biomol.Chem., 1, 2003
|
|
1ZC6
 
 | | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum. Northeast Structural Genomics Target Cvr23. | | Descriptor: | probable N-acetylglucosamine kinase | | Authors: | Vorobiev, S.M, Kuzin, A, Forouhar, F, Abashidze, M, Acton, T.B, Xiao, R, Ma, L.-C, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum
To be Published
|
|
1RNV
 
 | | REFINEMENT OF THE CRYSTAL STRUCTURE OF RIBONUCLEASE S. COMPARISON WITH AND BETWEEN THE VARIOUS RIBONUCLEASE A STRUCTURES | | Descriptor: | RIBONUCLEASE S, SULFATE ION | | Authors: | Kim, E.E, Varadarajan, R, Wyckoff, H.W, Richards, F.M. | | Deposit date: | 1992-02-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures.
Biochemistry, 31, 1992
|
|
2BIR
 
 | |
1Z6E
 
 | |
2BFY
 
 | | Complex of Aurora-B with INCENP and Hesperadin. | | Descriptor: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A, N-[2-OXO-3-((E)-PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]IMINO}METHYL)-2,6-DIHYDRO-1H-INDOL-5-YL]ETHANESULFONAMIDE | | Authors: | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | Deposit date: | 2004-12-15 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
2BN4
 
 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2005-03-18 | | Release date: | 2006-01-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
2BE2
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with R221239 | | Descriptor: | 4-(3,5-DIMETHYLPHENOXY)-5-(FURAN-2-YLMETHYLSULFANYLMETHYL)-3-IODO-6-METHYLPYRIDIN-2(1H)-ONE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Himmel, D.M, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Nguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
