8ERG
 
 | |
9MJM
 
 | | SOS1 IN COMPLEX WITH AN INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-({(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}amino)-3-(2-oxaspiro[3.3]heptan-6-yl)-5,6,7,8-tetrahydropyrido[4,3-d]pyrimidin-4(3H)-one, IMIDAZOLE, ... | | Authors: | Bell, J.A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploiting Solvent Exposed Salt-Bridge Interactions for the Discovery of Potent Inhibitors of SOS1 Using Free-Energy Perturbation Simulations.
Acs Med.Chem.Lett., 16, 2025
|
|
8KGA
 
 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
9MXQ
 
 | | Cryo-EM Structure of HIV-1 Reverse Transcriptase p66 Homodimer | | Descriptor: | Reverse transcriptase/ribonuclease H | | Authors: | Hollander, K, Devarkar, S.C, Tang, S, Ma, S, Xiong, Y, Anderson, K.S. | | Deposit date: | 2025-01-20 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Mechanistic basis for a novel dual-function Gag-Pol dimerizer potentiating CARD8 inflammasome activation and clearance of HIV-infected cells.
Npj Drug Discov, 2, 2025
|
|
4W9F
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 5) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | van Molle, I, Hewitt, S, Galdeano, C, Gadd, M.S, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
8ERF
 
 | |
9QFS
 
 | | Structure of CHIP E3 ubiquitin ligase TPR domain in complex with compound 8. | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, ~{N}-[(4~{R})-4-cyclohexyl-2,5-bis(oxidanylidene)imidazolidin-1-yl]-4,5,6,7-tetrakis(fluoranyl)-1~{H}-indole-3-carboxamide | | Authors: | Breed, J. | | Deposit date: | 2025-03-12 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Discovery of Small-Molecule Ligands for the E3 Ligase STUB1/CHIP from a DNA-Encoded Library Screen.
Acs Med.Chem.Lett., 16, 2025
|
|
7OUN
 
 | |
6NXC
 
 | |
4RC2
 
 | |
4RCH
 
 | | Discovery of 2-Pyridyl Ureas as Glucokinase Activators | | Descriptor: | 1-{3-[(2-ethylpyridin-3-yl)oxy]-5-(pyridin-2-ylsulfanyl)pyridin-2-yl}-3-methylurea, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W, Vigers, G.P.A. | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 2-pyridylureas as glucokinase activators.
J.Med.Chem., 57, 2014
|
|
6VKG
 
 | | Human carbonic anhydrase IX mimic with Epacadostat bound | | Descriptor: | N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, SULFATE ION, ZINC ION, ... | | Authors: | Peat, T.S, Carta, F, Supuran, C.T. | | Deposit date: | 2020-01-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray crystallography of Epacadostat in adduct with Carbonic Anhydrase IX.
Bioorg.Chem., 97, 2020
|
|
6VMX
 
 | | Structure of HD14 TCR in complex with HLA-B7 presenting an EBV epitope | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Farenc, C, Rossjohn, J, Gras, S. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Shared TCR Bias toward an Immunogenic EBV Epitope Dominates in HLA-B*07:02-Expressing Individuals.
J Immunol., 205, 2020
|
|
6VWS
 
 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
6O55
 
 | |
6W0W
 
 | | Structure of KHK in complex with compound 3 | | Descriptor: | 6-[(3~{R},4~{S})-3,4-bis(oxidanyl)pyrrolidin-1-yl]-2-[(2~{S},3~{R})-2-methyl-3-oxidanyl-azetidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, Ketohexokinase, SULFATE ION | | Authors: | Jasti, J. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of PF-06835919: A Potent Inhibitor of Ketohexokinase (KHK) for the Treatment of Metabolic Disorders Driven by the Overconsumption of Fructose.
J.Med.Chem., 63, 2020
|
|
7P5K
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-fluoranyl-3-[(2~{R})-2-propylpiperidin-1-yl]carbonyl-phenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
4R1C
 
 | |
6OIR
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 62 | | Descriptor: | 4-fluoro-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
7OZD
 
 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 34. | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-[6-(4-hydroxyphenyl)-1H-indazol-3-yl]benzamide, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-27 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
6W4Q
 
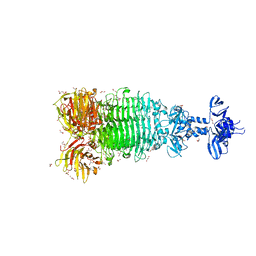 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
7OZF
 
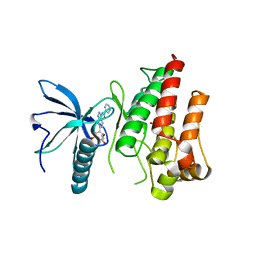 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 19. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Fibroblast growth factor receptor 1, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-28 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7OZY
 
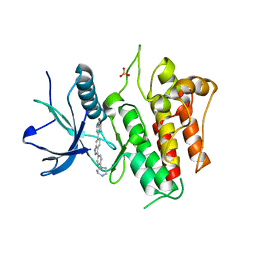 | | FGFR2 kinase domain (residues 461-763) in complex with 38. | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(4-piperazin-4-ium-1-ylphenyl)-1H-indazol-6-yl]phenol, Fibroblast growth factor receptor 2, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7QU9
 
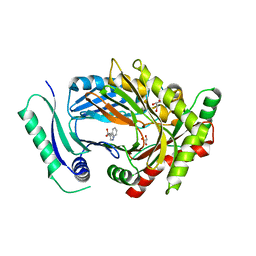 | | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii. | | Descriptor: | Anthranilate synthase component I family protein, GLYCEROL, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R, Back, C.B, Burton, N.B, Willis, C.L, Stach, J.E.M, Duke, P.W, Hawkins, C. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii.
To Be Published
|
|
4RG2
 
 | | Tudor Domain of Tumor suppressor p53BP1 with small molecule ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(tert-butylamino)propyl]benzamide, Tumor suppressor p53-binding protein 1, ... | | Authors: | Dong, A, Mader, P, James, L, Perfetti, M, Tempel, W, Frye, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1.
ACS Chem. Biol., 10, 2015
|
|
