2CAN
 
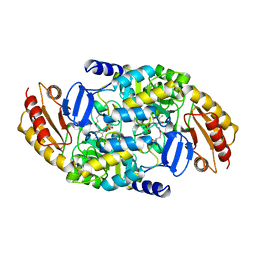 | |
1TJR
 
 | | Crystal structure of wild-type BX1 complexed with a sulfate ion | | Descriptor: | BX1, SULFATE ION | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2004-06-07 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
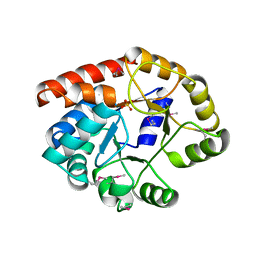
2CAG
 
 | | CATALASE COMPOUND II | | Descriptor: | CATALASE COMPOUND II, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gouet, P, Jouve, H.M, Hajdu, J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ferryl intermediates of catalase captured by time-resolved Weissenberg crystallography and UV-VIS spectroscopy.
Nat.Struct.Biol., 3, 1996
|
|
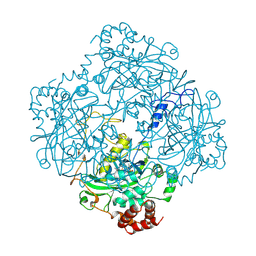
1TL1
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW451211 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFINYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
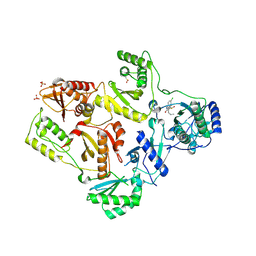
2CNO
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | Aza-peptide epoxide, Caspase-3 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Grutter, M.G. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
1ZML
 
 | | Crystal Structure of the Catalytic Domain of Factor XI in complex with (R)-1-(4-(4-(hydroxymethyl)-1,3,2-dioxaborolan-2-yl)phenethyl)guanidine | | Descriptor: | (R)-1-(4-(4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL)PHENETHYL)GUANIDINE, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M.J, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R.E, Strickler, J.E. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CNN
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EXPOXIDE, Caspase-3 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Grutter, M.G. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
2CDR
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EXPOXIDE, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Gruetter, M.G. | | Deposit date: | 2006-01-27 | | Release date: | 2007-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors.
Biochemistry, 45, 2006
|
|
2CNL
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EPOXIDE, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Grutter, M.G, Powers, J.C. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
1T7O
 
 | |
2CNX
 
 | | WDR5 and Histone H3 Lysine 4 dimethyl complex at 2.1 angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2CNK
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EXPOXIDE, CASPASE-3 P12 SUBUNIT, CASPASE-3 P17 SUBUNIT | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Grutter, M.G. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
1T21
 
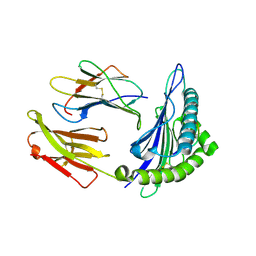 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9, monoclinic crystal | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
2CO0
 
 | | WDR5 and unmodified Histone H3 complex at 2.25 Angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
1T85
 
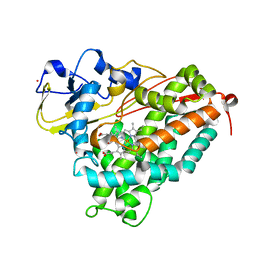 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam Mutant (L358P/C334A) | | Descriptor: | CAMPHOR, CARBON MONOXIDE, Cytochrome P450-cam, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1T8S
 
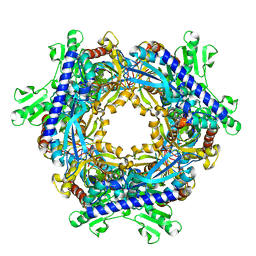 | |
2CGK
 
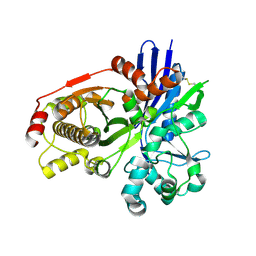 | |
2CEM
 
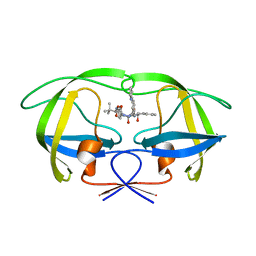 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
1Z8P
 
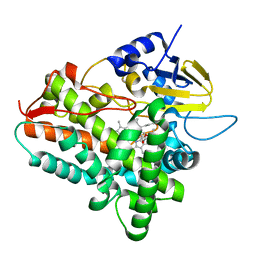 | | Ferrous dioxygen complex of the A245S cytochrome P450eryF | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, 6-deoxyerythronolide B hydroxylase, OXYGEN MOLECULE, ... | | Authors: | Poulos, T.L, Nagano, S. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF.
J.Biol.Chem., 280, 2005
|
|
2C77
 
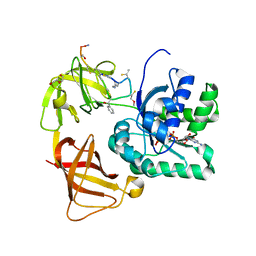 | | EF-Tu complexed with a GTP analog and the antibiotic GE2270 A | | Descriptor: | DI(HYDROXYETHYL)ETHER, ELONGATION FACTOR TU-B, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
1TAQ
 
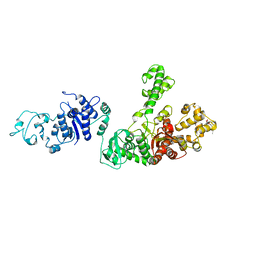 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
1ZP6
 
 | | Crystal Structure of Atu3015, a Putative Cytidylate Kinase from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR62 | | Descriptor: | SULFATE ION, hypothetical protein Atu3015 | | Authors: | Forouhar, F, Abashidze, M, Vorobiev, S.M, Kuzin, A, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Atu3015, a Putative Cytidylate Kinase from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR62
To be Published
|
|
1ZPB
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 4-Methyl-pentanoic acid {1-[4-guanidino-1-(thiazole-2-carbonyl)-butylcarbamoyl]-2-methyl-propyl}-amide | | Descriptor: | 4-METHYL-PENTANOIC ACID {1-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYLCARBAMOYL]-2-METHYL-PROPYL}-AMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
1TBW
 
 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
