1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BM2
 
 | |
1BM3
 
 | | IMMUNOGLOBULIN OPG2 FAB-PEPTIDE COMPLEX | | Descriptor: | IMMUNOGLOBULIN OPG2 FAB, CONSTANT DOMAIN, VARIABLE DOMAIN | | Authors: | Kodandapani, R, Veerapandian, L, Ni, C.Z, Chiou, C.-K, Whital, R, Kunicki, T.J, Ely, K.R. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational change in an anti-integrin antibody: structure of OPG2 Fab bound to a beta 3 peptide.
Biochem.Biophys.Res.Commun., 251, 1998
|
|
1BM4
 
 | | MOMLV CAPSID PROTEIN MAJOR HOMOLOGY REGION PEPTIDE ANALOG | | Descriptor: | PROTEIN (MOLONEY MURINE LEUKEMIA VIRUS CAPSID) | | Authors: | Clish, C.B, Peyton, D.H, Barklis, E. | | Deposit date: | 1998-07-28 | | Release date: | 1998-08-05 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human immunodeficiency virus type 1 (HIV-1) and moloney murine leukemia virus (MoMLV) capsid protein major-homology-region peptide analogs by NMR spectroscopy.
Eur.J.Biochem., 257, 1998
|
|
1BM5
 
 | |
1BM6
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Li, Y, Zhang, X, Melton, R, Ganu, V, Gonnella, N.C. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin-1 complexed to a potent, nonpeptidic inhibitor.
Biochemistry, 37, 1998
|
|
1BM7
 
 | |
1BM8
 
 | | DNA-BINDING DOMAIN OF MBP1 | | Descriptor: | TRANSCRIPTION FACTOR MBP1 | | Authors: | Xu, R.-M, Koch, C, Liu, Y, Horton, J.R, Knapp, D, Nasmyth, K, Cheng, X. | | Deposit date: | 1998-07-29 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis.
Structure, 5, 1997
|
|
1BM9
 
 | |
1BMA
 
 | | BENZYL METHYL AMINIMIDE INHIBITOR COMPLEXED TO PORCINE PANCREATIC ELASTASE | | Descriptor: | (1R)-1-benzyl-1-methyl-1-(2-{[4-(1-methylethyl)phenyl]amino}-2-oxoethyl)-2-{(2S)-4-methyl-2-[(trifluoroacetyl)amino]pentanoyl}diazanium, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Peisach, E, Casebier, D, Gallion, S.L, Furth, P, Petsko, G.A, Hogan Jr, J.C, Ringe, D. | | Deposit date: | 1995-05-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of a peptidomimetic aminimide inhibitor with elastase.
Science, 269, 1995
|
|
1BMB
 
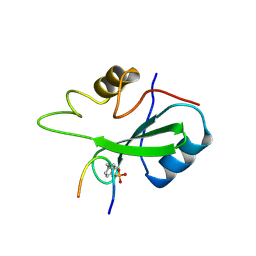 | | GRB2-SH2 DOMAIN IN COMPLEX WITH KPFY*VNVEF (PKF270-974) | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR BOUND PROTEIN 2), PROTEIN (PKF270-974) | | Authors: | Rondeau, J.M, Zurini, M. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
J.Med.Chem., 42, 1999
|
|
1BMC
 
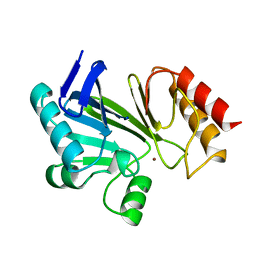 | | STRUCTURE OF A ZINC METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Carfi, A, Pares, S, Duee, E, Dideberg, O. | | Deposit date: | 1995-06-16 | | Release date: | 1996-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold.
EMBO J., 14, 1995
|
|
1BMD
 
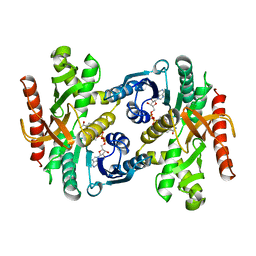 | |
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
1BMG
 
 | |
1BMK
 
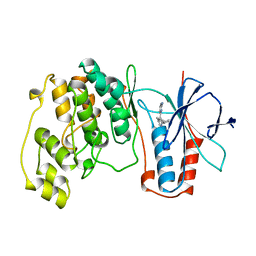 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BML
 
 | |
1BMM
 
 | |
1BMN
 
 | |
1BMO
 
 | | BM-40, FS/EC DOMAIN PAIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM-40, CALCIUM ION | | Authors: | Hohenester, E, Maurer, P, Timpl, R. | | Deposit date: | 1997-03-25 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a pair of follistatin-like and EF-hand calcium-binding domains in BM-40.
EMBO J., 16, 1997
|
|
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | Descriptor: | BONE MORPHOGENETIC PROTEIN-7 | | Authors: | Griffith, D.L, Scott, D.L. | | Deposit date: | 1995-12-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | Descriptor: | LQH III ALPHA-LIKE TOXIN | | Authors: | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
1BMS
 
 | |
1BMT
 
 | | HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE | | Descriptor: | CO-METHYLCOBALAMIN, METHIONINE SYNTHASE | | Authors: | Drennan, C.L, Huang, S, Drummond, J.T, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1994-09-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase.
Science, 266, 1994
|
|
