6TMK
 
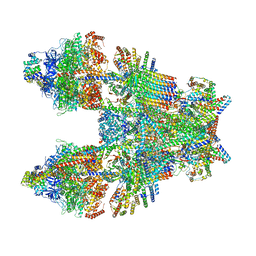 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, composite model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
7XKG
 
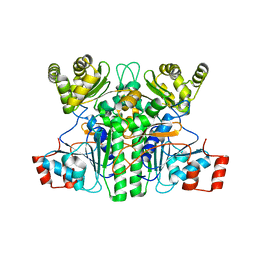 | | Crystal structure of an intramolecular mesacyl-CoA transferase from the 3-hydroxypropionic acid cycle of Roseiflexus castenholzii | | Descriptor: | Acyl-CoA transferase/carnitine dehydratase-like protein | | Authors: | Min, Z.Z, Fan, C.P, Wu, W.P, Xin, Y.Y, Liu, M.H, Zhang, X, Wang, Z.G, Xu, X.L. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Intramolecular Mesaconyl-Coenzyme A Transferase From the 3-Hydroxypropionic Acid Cycle of Roseiflexus castenholzii .
Front Microbiol, 13, 2022
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
6MS2
 
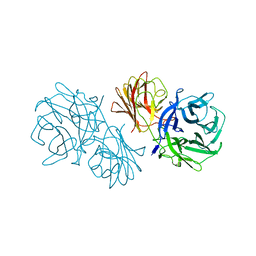 | | Crystal structure of the GH43 BlXynB protein from Bacillus licheniformis | | Descriptor: | CALCIUM ION, Glycoside Hydrolase Family 43 | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
7LZ9
 
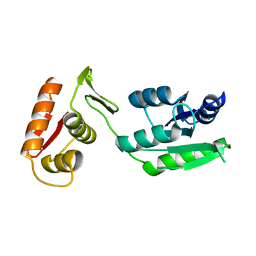 | | Inactive form of VanR from S. coelicolor | | Descriptor: | MAGNESIUM ION, Putative two-component system response regulator | | Authors: | Maciunas, L.J, Loll, P.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of full-length VanR from Streptomyces coelicolor in both the inactive and activated states.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LSA
 
 | | Ruminococcus bromii Amy12 with maltoheptaose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
6N7E
 
 | |
8AVU
 
 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the dimeric state | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8TLU
 
 | | E. coli MraY mutant-T23P | | Descriptor: | Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Li, Y.E, Clemons, W.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa.
Nat Microbiol, 9, 2024
|
|
8AVS
 
 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the presence of citrate and acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bacteriocin aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8AVT
 
 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the presence of glycerol 3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8AVR
 
 | | Racemic protein crystal structure of aureocin A53 from Staphylococcus aureus in the presence of sulfate | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin aureocin A53, D-Aureocin A53, ... | | Authors: | Lander, A.J, Baumann, P, Rizkallah, P, Jin, Y, Luk, L.Y.P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
6BZH
 
 | | Structure of mouse RIG-I tandem CARDs | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Babon, J.J, Nicholson, S.E. | | Deposit date: | 2017-12-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a second binding site on the TRIM25 B30.2 domain.
Biochem. J., 475, 2018
|
|
7LST
 
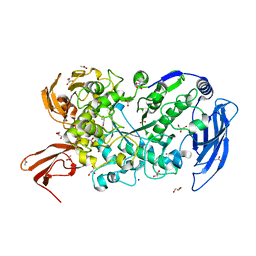 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
8AVB
 
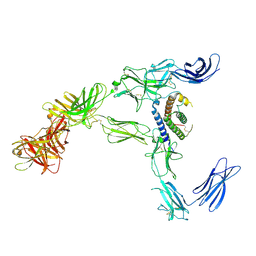 | | Cryo-EM structure for mouse leptin in complex with the mouse LEP-R ectodomain (1:2 mLEP:mLEPR model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6NV2
 
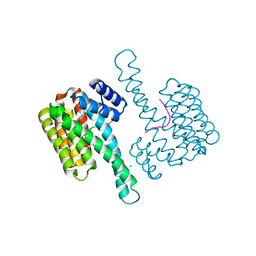 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
6HSD
 
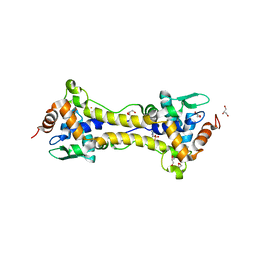 | | Crystal structure of the oxidized form of the transcription regulator RsrR | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-09-30 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
7LSU
 
 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
6C16
 
 | |
6VOR
 
 | | Crystal structure of macaque anti-HIV-1 antibody RM20E1 | | Descriptor: | GLYCINE, RM20E1 Fab heavy chain, RM20E1 Fab light chain | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
5ED3
 
 | | crystal structure of human Hint1 complexing with AP5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2015-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6SVE
 
 | | Protein allostery of the WW domain at atomic resolution: pCdc25C bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5J5F
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5EE2
 
 | |
