8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
1A8B
 
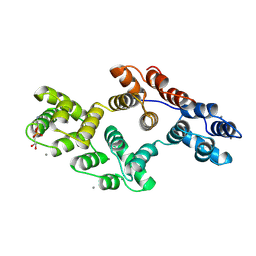 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOETHANOLAMINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLETHANOLAMINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
5UWP
 
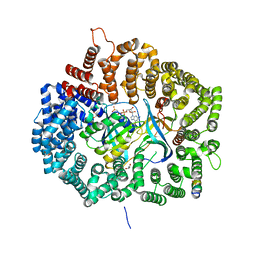 | |
8D7U
 
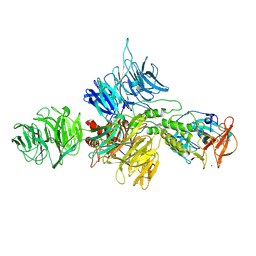 | |
8D81
 
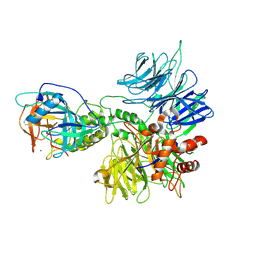 | | Cereblon~DDB1 bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7Y
 
 | |
5UWO
 
 | |
5UYZ
 
 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) mutant His147Arg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
8EYI
 
 | |
8EYK
 
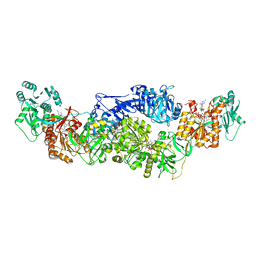 | |
8F93
 
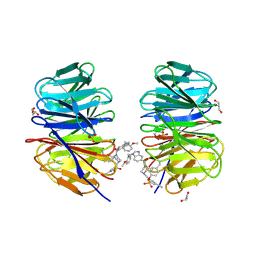 | | WDR5 covalently modified at Y228 by (R)-2-SF | | Descriptor: | 3-ethynyl-5-{[(3R)-4-{1-[(2-methoxyphenyl)methyl]-1H-benzimidazole-5-carbonyl}-3-methylpiperazin-1-yl]methyl}benzene-1-sulfonyl fluoride, CHLORIDE ION, GLYCEROL, ... | | Authors: | Taunton, J, Craven, G.B, Chen, Y. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes.
Nat.Chem., 15, 2023
|
|
5UWH
 
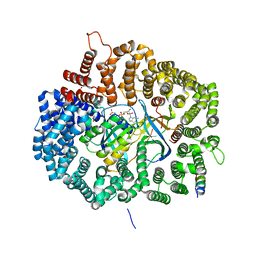 | |
5V67
 
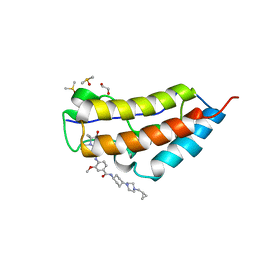 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH Volasertib | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | EMBER, S.W, ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-16 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 2021
|
|
5UYX
 
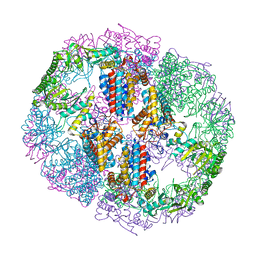 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
8EJM
 
 | |
5UGH
 
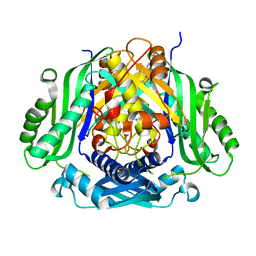 | | Crystal structure of Mat2a bound to the allosteric inhibitor PF-02929366 | | Descriptor: | 2-(7-chloro-5-phenyl[1,2,4]triazolo[4,3-a]quinolin-1-yl)-N,N-dimethylethan-1-amine, S-adenosylmethionine synthase isoform type-2 | | Authors: | Kaiser, S.E, Feng, J, Stewart, A.E. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A.
Nat. Chem. Biol., 13, 2017
|
|
8EHG
 
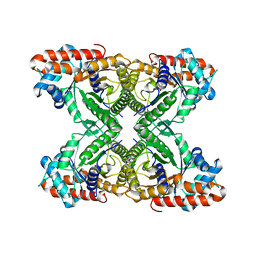 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
1ZKF
 
 | |
8EW2
 
 | |
1ZMZ
 
 | | Solution structure of the N-terminal domain (M1-S98) of human centrin 2 | | Descriptor: | Centrin-2 | | Authors: | Yang, A, Miron, S, Duchambon, P, Assairi, L, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-05-11 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly
Biochemistry, 45, 2006
|
|
8F1G
 
 | | Crystal structure of human WDR5 in complex with compound WM662 | | Descriptor: | (2S)-2-({(2S)-3-(3'-chloro[1,1'-biphenyl]-4-yl)-1-oxo-1-[(1H-tetrazol-5-yl)amino]propan-2-yl}oxy)propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, H. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of WDR5-MYC Interaction.
Acs Chem.Biol., 18, 2023
|
|
8EOM
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973 | | Descriptor: | 4-(4-methylpiperazine-1-sulfonyl)benzamide, SULFATE ION, TP53-binding protein 1, ... | | Authors: | The, J, Hong, Z, Headey, S, Gunzburg, M, Doak, B, James, L.I, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973
to be published
|
|
1AWR
 
 | | CYPA COMPLEXED WITH HAGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
8EAD
 
 | |
5UWT
 
 | |
