8UXU
 
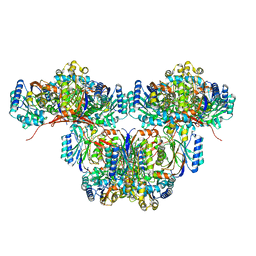 | | Cryo-EM structure of a bacterial nitrilase filament with a covalent adduct derived from benzonitrile hydrolysis | | Descriptor: | Nitrilase, benzaldehyde | | Authors: | Aguirre-Sampieri, S, Casanal, A, Emsley, P, Garza-Ramos, G. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of bacterial nitrilase reveals insight into oligomerization, substrate recognition, and catalysis.
J.Struct.Biol., 216, 2024
|
|
5HYG
 
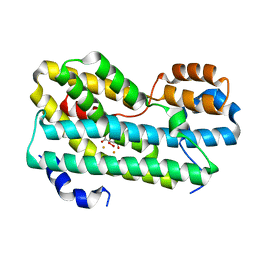 | | CmlI (peroxo bound state), arylamine oxygenase of chloramphenicol biosynthetic pathway | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, L(+)-TARTARIC ACID, ... | | Authors: | Knoot, C.J, Lipscomb, J.D. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of CmlI, the arylamine oxygenase from the chloramphenicol biosynthetic pathway.
J.Biol.Inorg.Chem., 21, 2016
|
|
5HYH
 
 | |
8VHV
 
 | |
8K1O
 
 | | mycobacterial efflux pump, AMPPNP bound state | | Descriptor: | CARDIOLIPIN, MAGNESIUM ION, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6C05
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA in relaxed state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
2M56
 
 | | The structure of the complex of cytochrome P450cam and its electron donor putidaredoxin determined by paramagnetic NMR spectroscopy | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hiruma, Y, Hass, M.A.S, Ubbink, M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
6F28
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2'-Amino-2'-carboxyethyl]-6-methyl-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.4A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
6FO6
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial inhibitor SCR0911 | | Descriptor: | 7-methoxy-3-methyl-2-[5-[4-(trifluoromethyloxy)phenyl]pyridin-3-yl]quinolin-4-ol, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6FO0
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial compound GSK932121 | | Descriptor: | 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, Chain I/V, Cytochrome b, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6FO2
 
 | | CryoEM structure of bovine cytochrome bc1 with no ligand bound | | Descriptor: | Cytochrome b, Cytochrome b-c1 complex subunit 1, mitochondrial, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
8P6P
 
 | | Mycoplasma pneumoniae small ribosomal subunit in chloramphenicol-treated cells | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-27 | | Release date: | 2024-11-20 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5I0V
 
 | |
5I0W
 
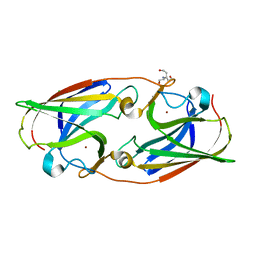 | |
8ONT
 
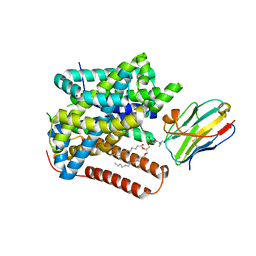 | | Structure of Setaria italica NRAT in complex with a nanobody | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, NRAMP related aluminium transporter, Nanobody1 | | Authors: | Ramanadane, K, Liziczai, M, Markovic, D, Straub, M.S, Rosalen, G.T, Udovcic, A, Dutzler, R, Manatschal, C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural and functional properties of a plant NRAMP-related aluminum transporter.
Elife, 12, 2023
|
|
5HDW
 
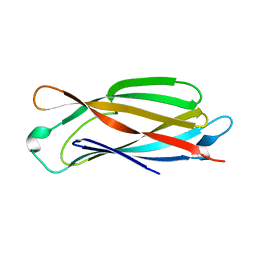 | |
7D7D
 
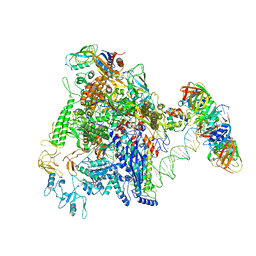 | | CryoEM structure of gp45-dependent transcription activation complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA polymerase clamp, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
2KA3
 
 | | Structure of EMILIN-1 C1Q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Corazza, A, Colebrooke, S.A, Cicero, D.O, Eliseo, T, Boyd, J, Doliana, R, Fogolari, F, Viglino, P, Colombatti, A, Campbell, I.D, Esposito, G. | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1
J.Biomol.Nmr, 43, 2009
|
|
2JLR
 
 | | Dengue virus 4 NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLV
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA and AMPPNP | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein
Embo J., 27, 2008
|
|
9F6P
 
 | |
6EEC
 
 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
6EDT
 
 | | Mycobacterium tuberculosis RNAP open promoter complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-10 | | Release date: | 2018-11-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
8XLA
 
 | | Mismatch Repair Complex | | Descriptor: | Beta sliding clamp, DNA mismatch repair protein MutL | | Authors: | Nirwal, S, Nair, D.T. | | Deposit date: | 2023-12-25 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the MutL-CTD:processivity-clamp complex provides insight regarding strand discrimination in non-methyl-directed DNA mismatch repair.
Nucleic Acids Res., 53, 2025
|
|
6GX7
 
 | | Tubulin-CopN-alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-26 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
