8WL5
 
 | | X-ray structure of Enterobacter cloacae allose-binding protein in free form | | Descriptor: | 1,2-ETHANEDIOL, Allose ABC transporter | | Authors: | Kamitori, S. | | Deposit date: | 2023-09-29 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structures of Enterobacter cloacae allose-binding protein in complexes with monosaccharides demonstrate its unique recognition mechanism for high affinity to allose.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
4N4G
 
 | |
8WLB
 
 | |
4N4S
 
 | | A Double Mutant Rat Erk2 in Complex With a Pyrazolo[3,4-d]pyrimidine Inhibitor | | Descriptor: | 3-[2-(benzyloxy)-8-methylquinolin-6-yl]-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Mitogen-activated protein kinase 1 | | Authors: | Hari, S.B, Maly, D.J, Merritt, E.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-Selective ATP-Competitive Inhibitors Control Regulatory Interactions and Noncatalytic Functions of Mitogen-Activated Protein Kinases.
Chem.Biol., 21, 2014
|
|
4N6W
 
 | | X-Ray Crystal Structure of Citrate-bound PhnZ | | Descriptor: | CITRATE ANION, FE (III) ION, Predicted HD phosphohydrolase PhnZ | | Authors: | Worsdorfer, B, Lingaraju, M, Yennawar, N.H, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8WWX
 
 | |
4N77
 
 | |
8WGL
 
 | | Crystal structure of Rhodothermus marinus substrate-binding protein (Hg soaking) | | Descriptor: | ABC-type uncharacterized transport system periplasmic component-like protein, MERCURY (II) ION | | Authors: | Nam, K.H. | | Deposit date: | 2023-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and bioinformatics analysis of single-domain substrate-binding protein from Rhodothermus marinus.
Biochem Biophys Rep, 37, 2024
|
|
8WMY
 
 | | Crystal structure of YqeK in complex with MG and ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Mo, X. | | Deposit date: | 2023-10-05 | | Release date: | 2023-10-25 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Mechanism of Ap4A Hydrolysis by YqeK of Staphylococcus pyogenes.
To be published
|
|
4V7S
 
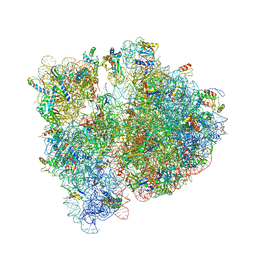 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4N84
 
 | | Crystal structure of 14-3-3zeta in complex with a 12-carbon-linker cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8WSW
 
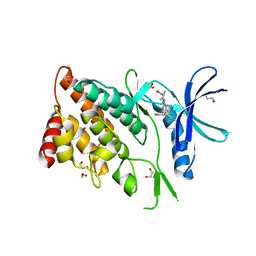 | | The Crystal Structure of LIMK2a from Biortus | | Descriptor: | 1,2-ETHANEDIOL, LIM domain kinase 2, ~{N}-[5-[2-[2,6-bis(chloranyl)phenyl]-5-[bis(fluoranyl)methyl]pyrazol-3-yl]-1,3-thiazol-2-yl]-2-methyl-propanamide | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of LIMK2a from Biortus.
To Be Published
|
|
8WTF
 
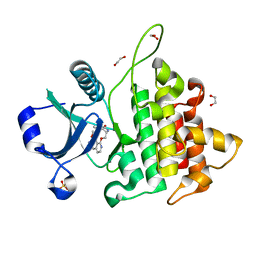 | | The Crystal Structure of IRAK4 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of IRAK4 from Biortus
To Be Published
|
|
4N9Q
 
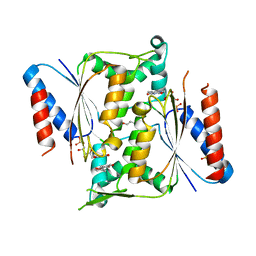 | | Crystal structure of paAzoR1 bound to ubiquinone-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Ryan, A, Kaplan, E, Crescente, V, Lowe, E, Preston, G.M, Sim, E. | | Deposit date: | 2013-10-21 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of NAD(P)H Quinone Oxidoreductase Activity in Azoreductases from P. aeruginosa: Azoreductases and NAD(P)H Quinone Oxidoreductases Belong to the Same FMN-Dependent Superfamily of Enzymes.
Plos One, 9, 2014
|
|
5WB8
 
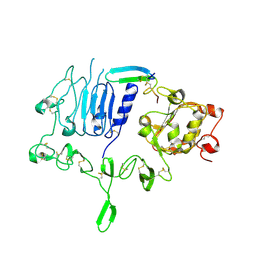 | | Crystal structure of the epidermal growth factor receptor extracellular region in complex with epigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Epigen, ... | | Authors: | Bessman, N.J, Freed, D.M, Moore, J.O, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Cell, 171, 2017
|
|
4NA4
 
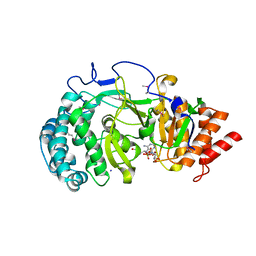 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4N94
 
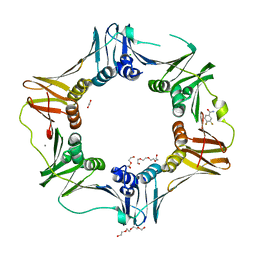 | | E. coli sliding clamp in complex with 3,4-difluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 3,4-difluorobenzamide, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
6SS8
 
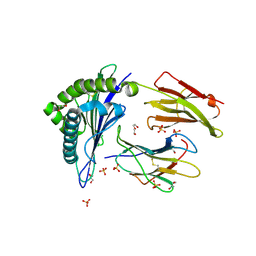 | | Human Leukocyte Antigen Class I A02 Carrying LLWNGPIAV | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Bovay, A. | | Deposit date: | 2019-09-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Identification of a superagonist variant of the immunodominant Yellow fever virus epitope NS4b214-222by combinatorial peptide library screening.
Mol.Immunol., 125, 2020
|
|
1DBG
 
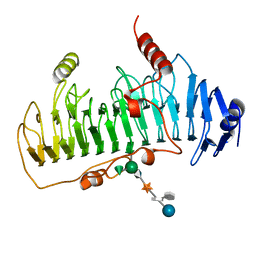 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
4NB1
 
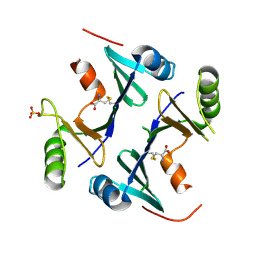 | | Crystal Structure of FosB from Staphylococcus aureus at 1.80 Angstrom Resolution with L-Cysteine-Cys9 Disulfide | | Descriptor: | CYSTEINE, Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4NBI
 
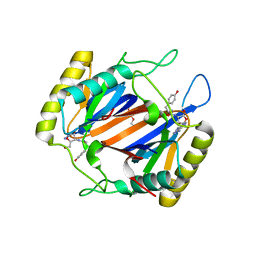 | | D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 1.86 Angstrom resolution | | Descriptor: | 3'-deoxy-3'-(D-tyrosylamino)adenosine, D-tyrosyl-tRNA(Tyr) deacylase, TRIETHYLENE GLYCOL | | Authors: | Ahmad, S, Routh, S.B, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanism of chiral proofreading during translation of the genetic code.
Elife, 2, 2013
|
|
4NBK
 
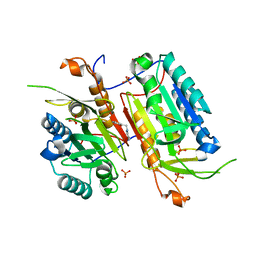 | |
4MLJ
 
 | |
6SX0
 
 | | Specific dsRNA recognition by wild type H7N1 NS1 RNA-binding domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Non-structural protein 1, ... | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
4MQI
 
 | |
