2WZS
 
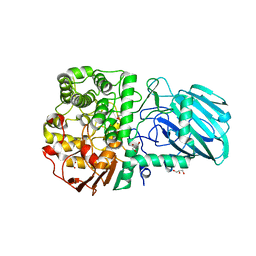 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhu, Y, Suits, M.D.L, Thompson, A, Chavan, S, Dinev, Z, Dumon, C, Smith, N, Moremen, K.W, Xiang, Y, Siriwardena, A, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
1NHZ
 
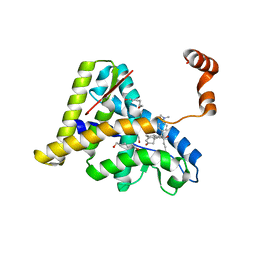 | | Crystal Structure of the Antagonist Form of Glucocorticoid Receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLUCOCORTICOID RECEPTOR, HEXANE-1,6-DIOL | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
6NAO
 
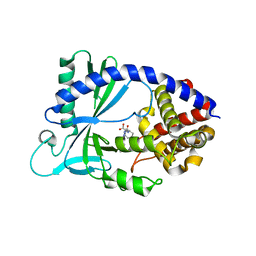 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, CYCLIC GMP-AMP SYNTHASE, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
2AAV
 
 | | Solution NMR structure of Filamin A domain 17 | | Descriptor: | Filamin A | | Authors: | Nakamura, F, Pudas, R, Heikkinen, O, Permi, P, Kilpelainen, I, Munday, A.D, Hartwig, J.H, Stossel, T.P, Ylanne, J. | | Deposit date: | 2005-07-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the GPIb-filamin A complex
Blood, 107, 2006
|
|
1THB
 
 | |
5DDK
 
 | | Succinyl-CoA:acetate CoA-transferase (AarCH6-N347A) in complex with CoA | | Descriptor: | Acetyl-CoA hydrolase, CHLORIDE ION, COENZYME A, ... | | Authors: | Kappock, T.J, Mullins, E.A, Murphy, J.R. | | Deposit date: | 2015-08-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Functional Dissection of the Bipartite Active Site of the Class I Coenzyme A (CoA)-Transferase Succinyl-CoA:Acetate CoA-Transferase.
Front Chem, 4, 2016
|
|
1UZA
 
 | | Crystallographic structure of a feruloyl esterase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FERULOYL ESTERASE A, SULFATE ION | | Authors: | McAuley, K.E, Svendsen, A, Patkar, S.A, Wilson, K.S. | | Deposit date: | 2004-03-05 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Feruloyl Esterase from Aspergillus Niger
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VC6
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutaion, cleaved in Imidazole and Mg2+ solutions | | Descriptor: | Hepatitis Delta virus ribozyme, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
4C3P
 
 | | Structure of dephosphorylated Aurora A (122-403) bound to TPX2 and AMPPCP | | Descriptor: | AURORA KINASE A, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Zorba, A, Kutter, S, Kern, D. | | Deposit date: | 2013-08-26 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular Mechanism of Aurora a Kinase Autophosphorylation and its Allosteric Activation by Tpx2.
Elife, 3, 2014
|
|
4PUZ
 
 | |
3NZ2
 
 | | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, ACETYL COENZYME *A, ... | | Authors: | Kim, Y, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor
To be Published
|
|
4C3R
 
 | | Structure of dephosphorylated Aurora A (122-403) bound to AMPPCP | | Descriptor: | AURORA KINASE A, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zorba, A, Kutter, S, Cho, Y.-J, Kern, D. | | Deposit date: | 2013-08-26 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular Mechanism of Aurora a Kinase Autophosphorylation and its Allosteric Activation by Tpx2.
Elife, 3, 2014
|
|
1RD6
 
 | | Crystal Structure of S. Marcescens Chitinase A Mutant W167A | | Descriptor: | Chitinase A | | Authors: | Aronson, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation.
Biosci.Biotechnol.Biochem., 70, 2006
|
|
3B8D
 
 | | Fructose 1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A, SULFATE ION | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2007-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
3B9D
 
 | | Crystal structure of Vibrio harveyi chitinase A complexed with pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Songsiriritthigul, C, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
4L7F
 
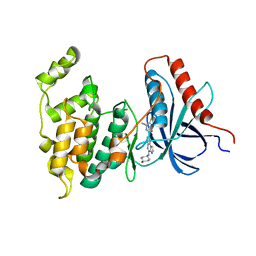 | | Co-crystal Structure of JNK1 and AX13587 | | Descriptor: | Mitogen-activated protein kinase 8, N-[1-(4-fluorophenyl)cyclopropyl]-4-[(trans-4-hydroxycyclohexyl)amino]imidazo[1,2-a]quinoxaline-8-carboxamide | | Authors: | Walter, R.L, Ranieri, G.M, Riggs, A.M, Weissig, H, Li, B, Shreder, K.R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hit-to-lead optimization and kinase selectivity of imidazo[1,2-a]quinoxalin-4-amine derived JNK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1JVT
 
 | |
4LUV
 
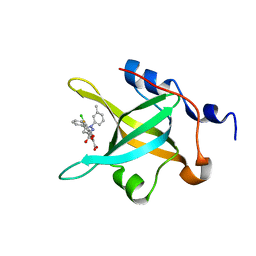 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 1-(3-methylphenyl)-5-phenyl-1H-pyrazole-3-carboxylic acid, 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
2CTB
 
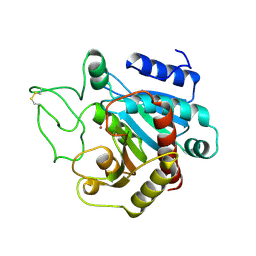 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CARBOXYPEPTIDASE A AND L-PHENYL LACTATE | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Teplyakov, A, Wilson, K.S, Orioli, P, Mangani, S. | | Deposit date: | 1993-04-02 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the complex between carboxypeptidase A and L-phenyl lactate.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4HLZ
 
 | | Crystal Structure of Fab C179 in Complex with a H2N2 influenza virus hemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-10-17 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a classical broadly neutralizing stem antibody in complex with a pandemic h2 influenza virus hemagglutinin.
J.Virol., 87, 2013
|
|
1JVU
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE A (COMPLEXED FORM) | | Descriptor: | CYTIDINE-2'-MONOPHOSPHATE, RIBONUCLEASE A | | Authors: | Vitagliano, L, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-08-31 | | Release date: | 2002-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Reversible Substrate-Induced Domain Motions in Ribonuclease A
Proteins, 46, 2002
|
|
1JS8
 
 | | Structure of a Functional Unit from Octopus Hemocyanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CU2-O2 CLUSTER, Hemocyanin, ... | | Authors: | Cuff, M.E, Miller, K.I, van Holde, K.E, Hendrickson, W.A. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a functional unit from Octopus hemocyanin.
J.Mol.Biol., 278, 1998
|
|
4E80
 
 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | Poly(A) RNA polymerase protein cid1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5RSA
 
 | |
1X2H
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
